---
title: LBT07
subtitle: Laboratory Test Results with Highest NCI CTCAE Grade Post-Baseline
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(tern)
library(dplyr)
library(forcats)
adsl <- random.cdisc.data::cadsl
adlb <- random.cdisc.data::cadlb
adlb_labels <- var_labels(adlb)
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
adsl <- df_explicit_na(adsl)
adlb <- df_explicit_na(adlb)
# Select worst post-baseline records.
adlb_f <- adlb %>%
filter(ATOXGR != "<Missing>") %>%
filter(ONTRTFL == "Y") %>%
filter(WGRLOFL == "Y" | WGRHIFL == "Y")
var_labels(adlb_f) <- adlb_labels
# Derive GRADE_DIR and GRADE_ANL to use in layout from ATOXGR
adlb_f <- adlb_f %>%
mutate(
GRADE_DIR = factor(
case_when(
ATOXGR %in% c("-1", "-2", "-3", "-4") & .data$WGRLOFL == "Y" ~ "LOW",
ATOXGR == "0" ~ "ZERO",
ATOXGR %in% c("1", "2", "3", "4") & .data$WGRHIFL == "Y" ~ "HIGH",
TRUE ~ "NONE"
),
levels = c("LOW", "ZERO", "HIGH", "NONE")
),
GRADE_ANL = forcats::fct_relevel(
forcats::fct_recode(ATOXGR,
`1` = "-1", `2` = "-2", `3` = "-3", `4` = "-4"
),
c("0", "1", "2", "3", "4")
)
) %>%
var_relabel(
GRADE_DIR = "Direction of Abnormality",
GRADE_ANL = "Analysis Grade"
)
# Construct analysis map
map <- expand.grid(
PARAM = levels(adlb$PARAM),
GRADE_DIR = c("LOW", "HIGH"),
GRADE_ANL = as.character(1:4),
stringsAsFactors = FALSE
) %>%
arrange(PARAM, desc(GRADE_DIR), GRADE_ANL)
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
:::: panel-tabset
## Standard Table
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result")}
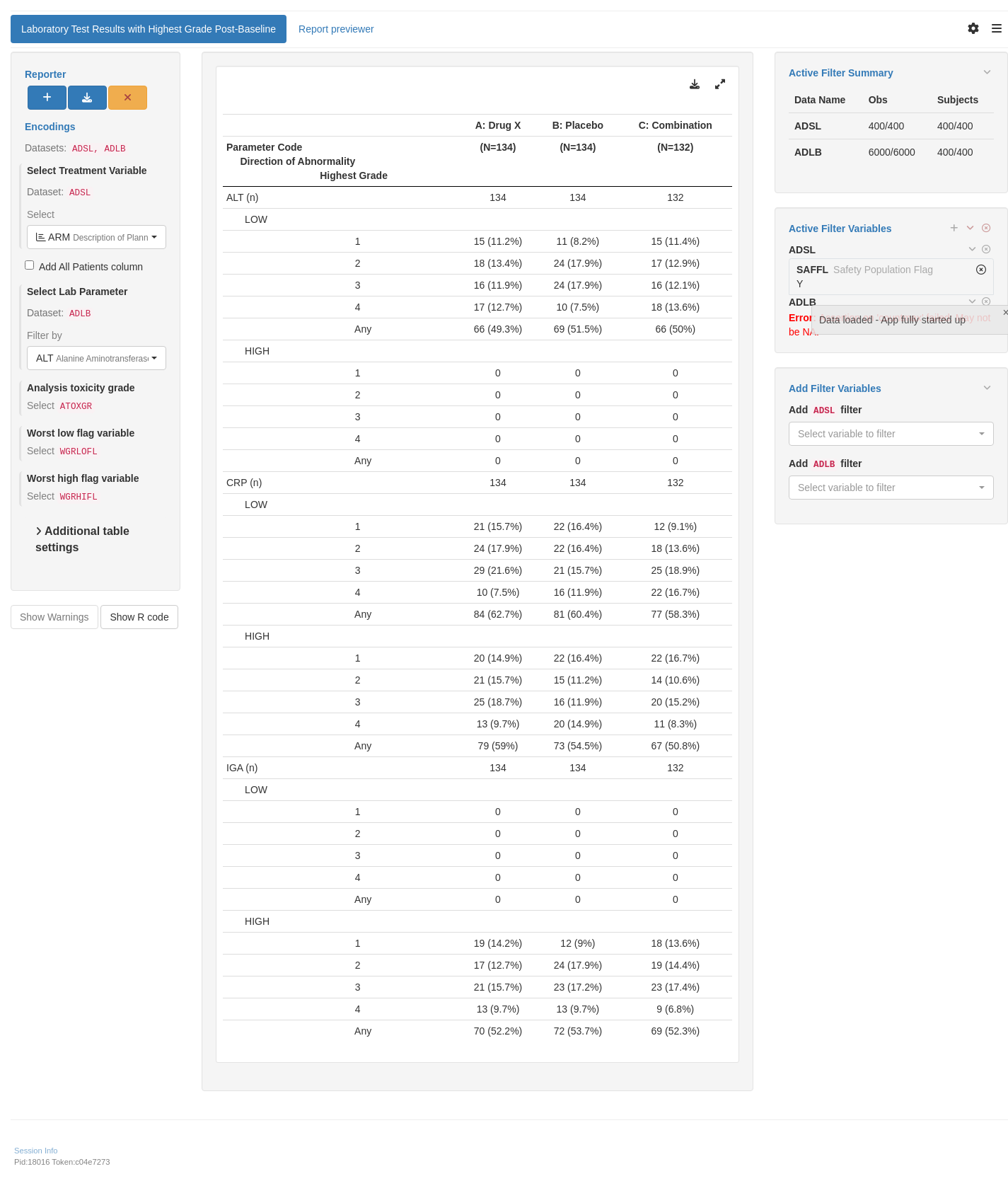
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
split_rows_by(
"PARAM",
label_pos = "topleft",
split_label = obj_label(adlb_f$PARAM)
) %>%
summarize_num_patients(
var = "USUBJID",
required = "ATOXGR",
.stats = "unique_count"
) %>%
split_rows_by(
"GRADE_DIR",
label_pos = "topleft",
split_label = obj_label(adlb_f$GRADE_DIR),
split_fun = trim_levels_to_map(map)
) %>%
count_abnormal_by_worst_grade(
var = "GRADE_ANL",
variables = list(id = "USUBJID", param = "PARAM", grade_dir = "GRADE_DIR"),
.indent_mods = 4L
) %>%
append_topleft(" Highest NCI CTCAE Grade")
result <- build_table(lyt = lyt, df = adlb_f, alt_counts_df = adsl)
result <- result %>% prune_table()
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
library(dplyr)
ADSL <- random.cdisc.data::cadsl
ADLB <- random.cdisc.data::cadlb %>%
filter(!AVISIT %in% c("SCREENING", "BASELINE"))
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADLB")]
## Reusable Configuration For Modules
ADSL <- data[["ADSL"]]
ADLB <- data[["ADLB"]]
## Setup App
app <- init(
data = data,
modules = modules(
tm_t_abnormality_by_worst_grade(
label = "Laboratory Test Results with Highest Grade Post-Baseline",
dataname = "ADLB",
arm_var = choices_selected(
choices = variable_choices(ADSL, subset = c("ARM", "ARMCD")),
selected = "ARM"
),
paramcd = choices_selected(
choices = value_choices(ADLB, "PARAMCD", "PARAM"),
selected = c("ALT", "CRP", "IGA")
),
add_total = FALSE
)
),
filter = (
teal_slices(
teal_slice("ADSL", "SAFFL", selected = "Y"),
teal_slice("ADLB", "ONTRTFL", selected = "Y")
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}