---
title: DMT01
subtitle: Demographics and Baseline Characteristics
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(tern)
library(dplyr)
library(tidyr)
adsl <- random.cdisc.data::cadsl
advs <- random.cdisc.data::cadvs
adsub <- random.cdisc.data::cadsub
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
adsl <- df_explicit_na(adsl)
advs <- df_explicit_na(advs)
adsub <- df_explicit_na(adsub)
# Change description in variable SEX.
adsl <- adsl %>%
mutate(
SEX = factor(case_when(
SEX == "M" ~ "Male",
SEX == "F" ~ "Female",
SEX == "U" ~ "Unknown",
SEX == "UNDIFFERENTIATED" ~ "Undifferentiated"
)),
AGEGR1 = factor(
case_when(
between(AGE, 18, 40) ~ "18-40",
between(AGE, 41, 64) ~ "41-64",
AGE > 64 ~ ">=65"
),
levels = c("18-40", "41-64", ">=65")
),
BMRKR1_CAT = factor(
case_when(
BMRKR1 < 3.5 ~ "LOW",
BMRKR1 >= 3.5 & BMRKR1 < 10 ~ "MEDIUM",
BMRKR1 >= 10 ~ "HIGH"
),
levels = c("LOW", "MEDIUM", "HIGH")
)
) %>%
var_relabel(
BMRKR1_CAT = "Biomarker 1 Categories"
)
# The developer needs to do pre-processing to add necessary variables based on ADVS to analysis dataset.
# Obtain SBP, DBP and weight.
get_param_advs <- function(pname, plabel) {
ds <- advs %>%
filter(PARAM == plabel & AVISIT == "BASELINE") %>%
select(USUBJID, AVAL)
colnames(ds) <- c("USUBJID", pname)
ds
}
# The developer needs to do pre-processing to add necessary variables based on ADSUB to analysis dataset.
# Obtain baseline BMI (BBMISI).
get_param_adsub <- function(pname, plabel) {
ds <- adsub %>%
filter(PARAM == plabel) %>%
select(USUBJID, AVAL)
colnames(ds) <- c("USUBJID", pname)
ds
}
adsl <- adsl %>%
left_join(get_param_advs("SBP", "Systolic Blood Pressure"), by = "USUBJID") %>%
left_join(get_param_advs("DBP", "Diastolic Blood Pressure"), by = "USUBJID") %>%
left_join(get_param_advs("WGT", "Weight"), by = "USUBJID") %>%
left_join(get_param_adsub("BBMISI", "Baseline BMI"), by = "USUBJID")
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
:::::::: panel-tabset
## Table with an Additional <br/> Study-Specific Continuous Variable
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result")}
vars <- c("AGE", "AGEGR1", "SEX", "ETHNIC", "RACE", "BMRKR1")
var_labels <- c(
"Age (yr)",
"Age Group",
"Sex",
"Ethnicity",
"Race",
"Continous Level Biomarker 1"
)
result <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
add_overall_col("All Patients") %>%
analyze_vars(
vars = vars,
var_labels = var_labels
) %>%
build_table(adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with an Additional <br/> Study-Specific Categorical Variable
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant2, test = list(result_v2 = "result")}
vars <- c("AGE", "AGEGR1", "SEX", "ETHNIC", "RACE", "BMRKR1_CAT")
var_labels <- c(
"Age (yr)",
"Age Group",
"Sex",
"Ethnicity",
"Race",
"Biomarker 1 Categories"
)
result <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
analyze_vars(
vars = vars,
var_labels = var_labels
) %>%
build_table(adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Subgrouping <br/> for Some Analyses
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant3, test = list(result_v3 = "result")}
split_fun <- drop_split_levels
result <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
analyze_vars(
vars = c("AGE", "SEX", "RACE"),
var_labels = c("Age", "Sex", "Race")
) %>%
split_rows_by("STRATA1",
split_fun = split_fun
) %>%
analyze_vars("BMRKR1") %>%
build_table(adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant3")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Additional Vital <br/> Signs Baseline Values
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant4, test = list(result_v4 = "result")}
result <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
analyze_vars(
vars = c("AGE", "SEX", "RACE", "DBP", "SBP"),
var_labels = c(
"Age (yr)",
"Sex",
"Race",
"Diastolic Blood Pressure",
"Systolic Blood Pressure"
)
) %>%
build_table(adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant4")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Additional <br/> Values from ADSUB
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant5, test = list(result_v5 = "result")}
result <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
analyze_vars(
vars = c("AGE", "SEX", "RACE", "BBMISI"),
var_labels = c(
"Age (yr)",
"Sex",
"Race",
"Baseline BMI"
)
) %>%
build_table(adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant5")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
::::::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
ADSL <- random.cdisc.data::cadsl
# Include `EOSDY` and `DCSREAS` variables below because they contain missing data.
stopifnot(
any(is.na(ADSL$EOSDY)),
any(is.na(ADSL$DCSREAS))
)
})
join_keys(data) <- default_cdisc_join_keys["ADSL"]
## Setup App
app <- init(
data = data,
modules = modules(
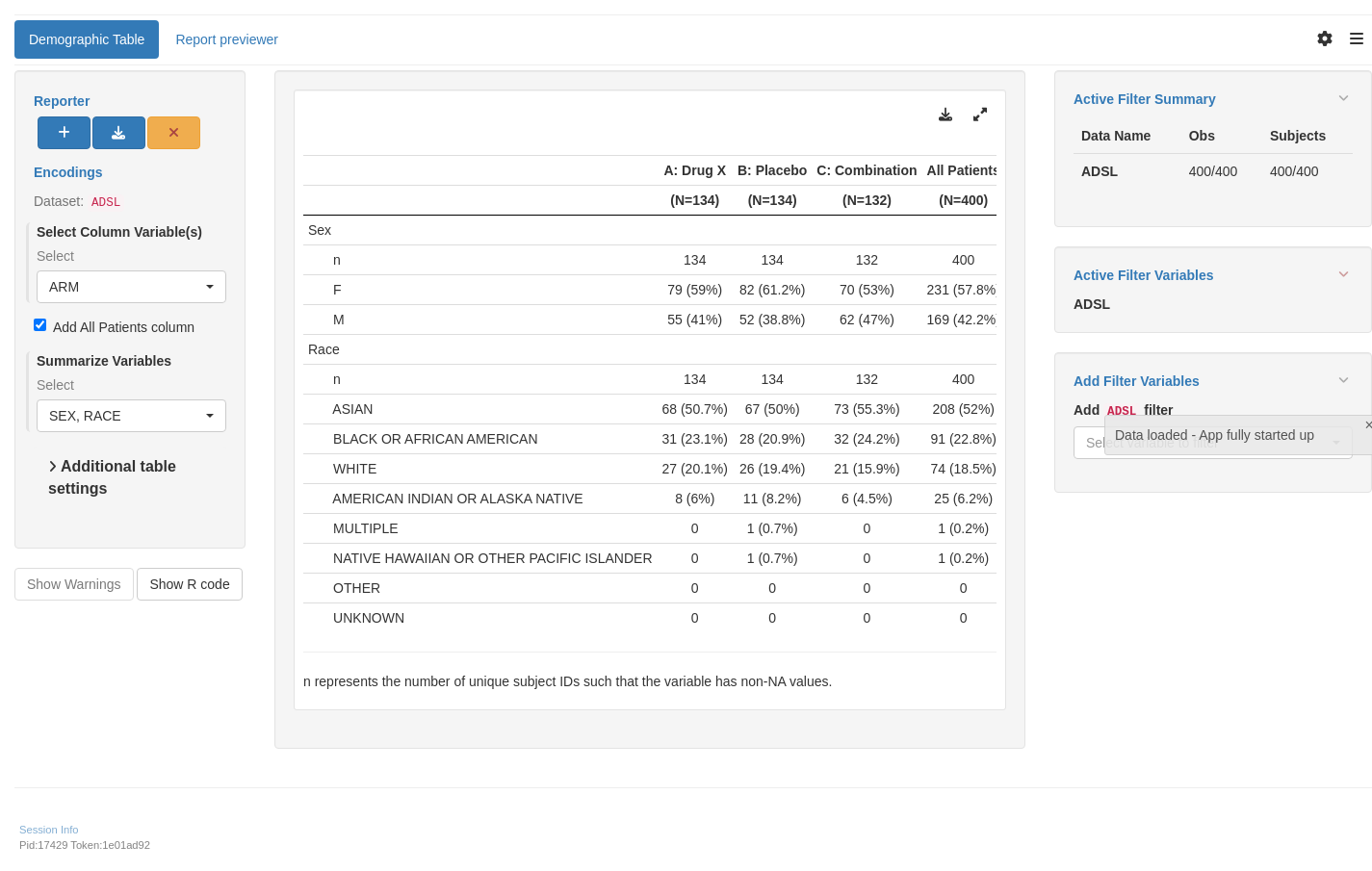
tm_t_summary(
label = "Demographic Table",
dataname = "ADSL",
arm_var = choices_selected(c("ARM", "ARMCD"), "ARM"),
summarize_vars = choices_selected(
c("SEX", "RACE", "BMRKR2", "EOSDY", "DCSREAS"),
c("SEX", "RACE")
),
useNA = "ifany"
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}