---
title: DORT01
subtitle: Duration of Response
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(tern)
library(dplyr)
adsl <- random.cdisc.data::cadsl
adtte <- random.cdisc.data::cadtte
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
adsl <- df_explicit_na(adsl)
adtte <- df_explicit_na(adtte)
adtte_f <- adtte %>%
filter(PARAMCD == "CRSD" & BMEASIFL == "Y") %>%
dplyr::mutate(
AVAL = day2month(AVAL),
is_event = CNSR == 0,
is_not_event = CNSR == 1,
EVNT1 = factor(
case_when(
is_event == TRUE ~ "Responders with subsequent event (%)",
is_event == FALSE ~ "Responders without subsequent event (%)"
),
levels = c("Responders with subsequent event (%)", "Responders without subsequent event (%)")
),
EVNTDESC = factor(EVNTDESC)
)
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
::::::: panel-tabset
## Standard Table
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ARM", ref_group = "A: Drug X") %>%
count_values(
vars = "USUBJID",
values = unique(adtte$USUBJID),
.labels = c(count = "Responders"),
.stats = "count"
) %>%
analyze_vars(
vars = "is_event",
.stats = "count_fraction",
.labels = c(count_fraction = "Responders with subsequent event (%)"),
.indent_mods = c(count_fraction = 1L),
show_labels = "hidden",
) %>%
split_rows_by(
"EVNT1",
split_label = "Earliest contributing event",
split_fun = keep_split_levels("Responders with subsequent event (%)"),
label_pos = "visible",
child_labels = "hidden",
indent_mod = 2L,
) %>%
analyze("EVNTDESC") %>%
analyze_vars(
vars = "is_not_event",
.stats = "count_fraction",
.labels = c(count_fraction = "Responders without subsequent event (%)"),
.indent_mods = c(count_fraction = 1L),
nested = FALSE,
show_labels = "hidden"
) %>%
surv_time(
vars = "AVAL",
var_labels = "Duration of response (Months)",
is_event = "is_event"
) %>%
surv_timepoint(
vars = "AVAL",
var_labels = "Months duration",
is_event = "is_event",
time_point = 12
)
result <- build_table(lyt, df = adtte_f, alt_counts_df = adsl) %>%
prune_table(prune_func = prune_zeros_only)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table Selecting <br/> Sections to Display
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant2, test = list(result_v2 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ARM", ref_group = "A: Drug X") %>%
count_values(
vars = "USUBJID",
values = unique(adtte$USUBJID),
.labels = c(count = "Responders"),
.stats = "count"
) %>%
analyze_vars(
vars = "is_event",
.stats = "count_fraction",
.labels = c(count_fraction = "Responders with subsequent event (%)"),
.indent_mods = c(count_fraction = 1L),
show_labels = "hidden",
) %>%
split_rows_by(
"EVNT1",
split_label = "Earliest contributing event",
split_fun = keep_split_levels("Responders with subsequent event (%)"),
label_pos = "visible",
child_labels = "hidden",
indent_mod = 2L,
) %>%
analyze("EVNTDESC") %>%
analyze_vars(
vars = "is_not_event",
.stats = "count_fraction",
.labels = c(count_fraction = "Responders without subsequent event (%)"),
.indent_mods = c(count_fraction = 1L),
nested = FALSE,
show_labels = "hidden"
) %>%
surv_time(
vars = "AVAL",
var_labels = "Duration of response (Months)",
is_event = "is_event",
table_names = "surv_time"
) %>%
coxph_pairwise(
vars = "AVAL",
is_event = "is_event",
var_labels = c("Unstratified Analysis"),
control = control_coxph(pval_method = "log-rank"),
table_names = "cox_pair"
)
result <- build_table(lyt, df = adtte_f, alt_counts_df = adsl) %>%
prune_table(prune_func = prune_zeros_only)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table Modifying Analysis Details <br/> like Conf. Type and Alpha Level
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant3, test = list(result_v3 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ARM", ref_group = "A: Drug X") %>%
count_values(
vars = "USUBJID",
values = unique(adtte$USUBJID),
.labels = c(count = "Responders"),
.stats = "count"
) %>%
analyze_vars(
vars = "is_event",
.stats = "count_fraction",
.labels = c(count_fraction = "Responders with subsequent event (%)"),
.indent_mods = c(count_fraction = 1L),
show_labels = "hidden",
) %>%
split_rows_by(
"EVNT1",
split_label = "Earliest contributing event",
split_fun = keep_split_levels("Responders with subsequent event (%)"),
label_pos = "visible",
child_labels = "hidden",
indent_mod = 2L,
) %>%
analyze("EVNTDESC") %>%
analyze_vars(
vars = "is_not_event",
.stats = "count_fraction",
.labels = c(count_fraction = "Responders without subsequent event (%)"),
.indent_mods = c(count_fraction = 1L),
nested = FALSE,
show_labels = "hidden"
) %>%
surv_time(
vars = "AVAL",
var_labels = "Duration of response (Months)",
is_event = "is_event",
control = control_surv_time(conf_level = 0.90, conf_type = "log-log")
) %>%
surv_timepoint(
vars = "AVAL",
var_labels = "Months duration",
is_event = "is_event",
time_point = 12,
control = control_surv_timepoint(conf_level = 0.975)
)
result <- build_table(lyt, df = adtte_f, alt_counts_df = adsl) %>%
prune_table(prune_func = prune_zeros_only)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant3")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table Modifying Time Point for <br/> the "XX Months duration" Analysis
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant4, test = list(result_v4 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ARM", ref_group = "A: Drug X") %>%
count_values(
vars = "USUBJID",
values = unique(adtte$USUBJID),
.labels = c(count = "Responders"),
.stats = "count"
) %>%
analyze_vars(
vars = "is_event",
.stats = "count_fraction",
.labels = c(count_fraction = "Responders with subsequent event (%)"),
.indent_mods = c(count_fraction = 1L),
show_labels = "hidden",
) %>%
split_rows_by(
"EVNT1",
split_label = "Earliest contributing event",
split_fun = keep_split_levels("Responders with subsequent event (%)"),
label_pos = "visible",
child_labels = "hidden",
indent_mod = 2L,
) %>%
analyze("EVNTDESC") %>%
analyze_vars(
vars = "is_not_event",
.stats = "count_fraction",
.labels = c(count_fraction = "Responders without subsequent event (%)"),
.indent_mods = c(count_fraction = 1L),
nested = FALSE,
show_labels = "hidden"
) %>%
surv_time(
vars = "AVAL",
var_labels = "Duration of response (Months)",
is_event = "is_event"
) %>%
surv_timepoint(
vars = "AVAL",
var_labels = "Months duration",
is_event = "is_event",
time_point = 6
)
result <- build_table(lyt, df = adtte_f, alt_counts_df = adsl) %>%
prune_table(prune_func = prune_zeros_only)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant4")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
:::::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
ADSL <- random.cdisc.data::cadsl
ADTTE <- random.cdisc.data::cadtte
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
ADSL <- df_explicit_na(ADSL)
ADTTE <- df_explicit_na(ADTTE)
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADTTE")]
## Reusable Configuration For Modules
ADSL <- data[["ADSL"]]
ADTTE <- data[["ADTTE"]]
arm_ref_comp <- list(
ACTARMCD = list(
ref = "ARM B",
comp = c("ARM A", "ARM C")
),
ARM = list(
ref = "B: Placebo",
comp = c("A: Drug X", "C: Combination")
)
)
## Setup App
app <- init(
data = data,
modules = modules(
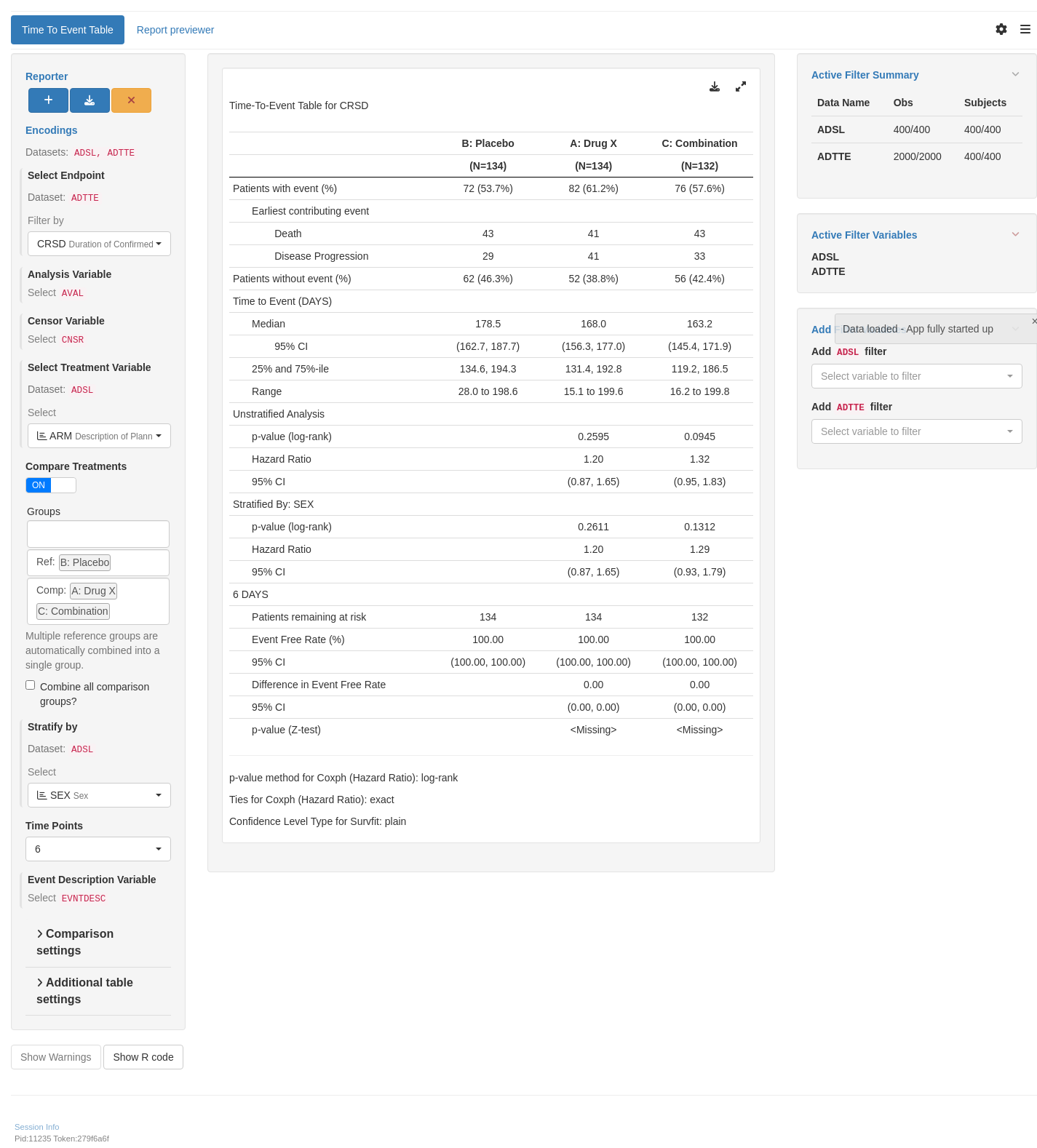
tm_t_tte(
label = "Time To Event Table",
dataname = "ADTTE",
arm_var = choices_selected(
variable_choices(ADSL, c("ARM", "ARMCD", "ACTARMCD")),
"ARM"
),
arm_ref_comp = arm_ref_comp,
paramcd = choices_selected(
value_choices(ADTTE, "PARAMCD", "PARAM"),
"CRSD"
),
strata_var = choices_selected(
variable_choices(ADSL, c("SEX", "BMRKR2")),
"SEX"
),
time_points = choices_selected(c(6, 8), 6),
event_desc_var = choices_selected(
variable_choices(ADTTE, "EVNTDESC"),
"EVNTDESC",
fixed = TRUE
)
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}