---
title: COXT02
subtitle: Multivariable Cox Regression
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(dplyr)
library(tern)
library(broom)
adtte <- random.cdisc.data::cadtte
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
adtte <- df_explicit_na(adtte)
anl <- adtte %>%
filter(
PARAMCD == "OS",
SEX %in% c("F", "M"),
RACE %in% c("ASIAN", "BLACK OR AFRICAN AMERICAN", "WHITE")
) %>%
mutate(
ARM = droplevels(relevel(ARM, "B: Placebo")),
SEX = droplevels(SEX),
RACE = droplevels(RACE)
) %>%
mutate(EVENT = 1 - CNSR) %>%
var_relabel(
ARM = "Planned Arm",
SEX = "Sex",
RACE = "Race",
AGE = "Age"
)
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
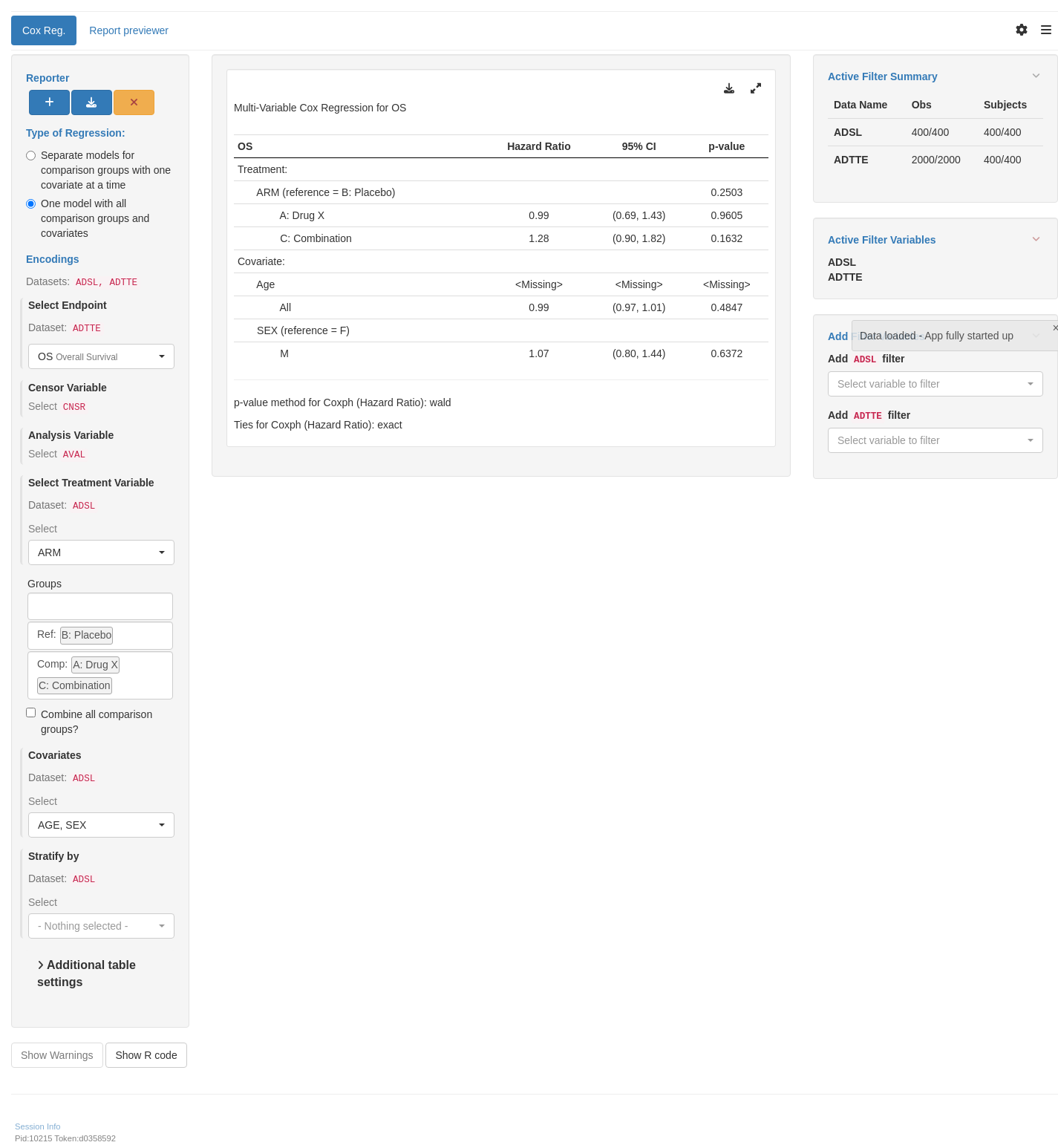
Analysis based on multivariable Cox models is usually not performed for the Clinical Study Report (CSR) or regulatory documents, serving exploratory purposes only (e.g. for publication). In practice, the model usually includes only the main effects (without interaction terms). It produces the estimates for each of the covariates included in the model. The analysis follows the same principles (i.e. stratified vs. unstratified analysis and tie handling) as the general Cox model analysis also used in `COXT01`. Since there is usually no pre-specified hypothesis testing for such analysis, the p-values must be interpreted with caution.
::::: panel-tabset
## Multivariable Cox Regression
The `summarize_coxreg` function fits, tidies and arranges a Cox regression model in a table layout using the `rtables` framework. For a multivariable Cox regression model, argument `multivar` must be set to `TRUE`. Arguments `variables` and `control` can be specified to set up the model (see `?summarize_coxreg` for more details and customization options). All variables specified within `variables` must be present in the data used when building the table.
To see the same model as a `data.frame` object, these two arguments (as well as the data) can be passed to the `fit_coxreg_multivar` function, and the resulting list tidied using `broom::tidy()`.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result")}
variables <- list(
time = "AVAL",
event = "EVENT",
arm = "ARM",
covariates = c("SEX", "AGE")
)
lyt <- basic_table() %>%
summarize_coxreg(variables = variables, multivar = TRUE) %>%
append_topleft("Effect/Covariate Included in the Model")
result <- build_table(lyt = lyt, df = anl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Multivariable Cox Regression <br/> with Interaction Term
The estimation of interaction terms is not supported.
Interaction terms are not included in the GDSR. For this reason and because we must take precautions when fitting such models, this functionality has not been translated in `fit_coxreg_multivar`. Please remove interaction terms or, if required by the study, refer to the `survival::coxph` function. Aside from this, using `tern` the developer must add the necessary variables to the analysis dataset during pre-processing based on ADVS or ADSUB. An example can be found in `DMT01`.
## Multivariable Cox Regression <br/> Specifying Covariates
This option is not supported.
See the *Multivariable Cox Regression with Interaction Term* tab for more details.
## Multivariable Cox Regression <br/> Specifying Covariates from ADSUB
This option is not supported.
See the *Multivariable Cox Regression with Interaction Term* tab for more details.
## Multivariable Cox Regression <br/> Setting Strata, Ties, Alpha Level, Statistics
Additional controls can be customized using `control_coxreg` (see `?control_coxreg`) such as the ties calculation method and the confidence level. Stratification variables can be added via the `strata` element of the `variables` list.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant2, test = list(result_v2 = "result")}
variables <- list(
time = "AVAL",
event = "EVENT",
arm = "ARMCD",
covariates = c("SEX", "AGE"),
strata = "RACE"
)
control <- control_coxreg(
conf_level = 0.9,
ties = "efron"
)
lyt <- basic_table() %>%
summarize_coxreg(
variables = variables,
control = control,
multivar = TRUE,
.stats = c("hr", "ci")
) %>%
append_topleft("Effect/Covariate Included in the Model")
result <- build_table(lyt = lyt, df = anl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Multivariable Cox Regression <br/> with Selection Process for Covariates
See the *Multivariable Cox Regression with Interaction Term* tab.
## Data Setup
```{r setup}
#| code-fold: show
```
:::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
ADSL <- random.cdisc.data::cadsl
ADTTE <- random.cdisc.data::cadtte
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADTTE")]
## Reusable Configuration For Modules
ADTTE <- data[["ADTTE"]]
arm_ref_comp <- list(
ACTARMCD = list(
ref = "ARM B",
comp = c("ARM A", "ARM C")
),
ARM = list(
ref = "B: Placebo",
comp = c("A: Drug X", "C: Combination")
)
)
## Setup App
app <- init(
data = data,
modules = modules(
tm_t_coxreg(
label = "Cox Reg.",
dataname = "ADTTE",
arm_var = choices_selected(c("ARM", "ARMCD", "ACTARMCD"), "ARM"),
arm_ref_comp = arm_ref_comp,
paramcd = choices_selected(
value_choices(ADTTE, "PARAMCD", "PARAM"), "OS"
),
strata_var = choices_selected(
c("SEX", "STRATA1", "STRATA2"), NULL
),
cov_var = choices_selected(
c("AGE", "SEX", "RACE"), c("AGE", "SEX")
),
multivariate = TRUE
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}