---
title: AET01_AESI
subtitle: Safety Summary (Adverse Events of Special Interest)
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(tern)
library(dplyr)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
adsl <- filter(adsl, SAFFL == "Y")
adae <- filter(adae, ANL01FL == "Y" & SAFFL == "Y")
adsl <- df_explicit_na(adsl)
adae <- df_explicit_na(adae)
not_resolved <- adae %>%
filter(!(AEOUT %in% c("RECOVERED/RESOLVED", "FATAL", "RECOVERED/RESOLVED WITH SEQUELAE"))) %>%
distinct(USUBJID) %>%
mutate(NOT_RESOLVED = "Y")
adae <- adae %>%
left_join(not_resolved, by = c("USUBJID")) %>%
mutate(
ALL_RESOLVED = with_label(
is.na(NOT_RESOLVED),
"Total number of patients with all non-fatal AEs resolved"
),
NOT_RESOLVED = with_label(
!is.na(NOT_RESOLVED),
"Total number of patients with at least one unresolved or ongoing non-fatal AE"
)
)
adae <- adae %>%
mutate(
AEDECOD = as.character(AEDECOD),
WD = with_label(
AEACN == "DRUG WITHDRAWN",
"Total number of patients with study drug withdrawn due to AE"
),
DSM = with_label(
AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"Total number of patients with dose modified/interrupted due to AE"
),
CONTRT = with_label(
AECONTRT == "Y",
"Total number of patients with treatment received for AE"
),
SER = with_label(
AESER == "Y",
"Total number of patients with at least one serious AE"
),
REL = with_label(
AEREL == "Y",
"Total number of patients with at least one related AE"
),
ALL_RESOLVED_WD = with_label(
WD == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with study drug withdrawn due to resolved AE"
),
ALL_RESOLVED_DSM = with_label(
DSM == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with dose modified/interrupted due to resolved AE"
),
ALL_RESOLVED_CONTRT = with_label(
CONTRT == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with treatment received for resolved AE"
),
NOT_RESOLVED_WD = with_label(
WD == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with study drug withdrawn due to unresolved or ongoing AE"
),
NOT_RESOLVED_DSM = with_label(
DSM == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with dose modified/interrupted due to unresolved or ongoing AE"
),
NOT_RESOLVED_CONTRT = with_label(
CONTRT == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with treatment received for unresolved or ongoing AE"
),
SERWD = with_label(
AESER == "Y" & AEACN == "DRUG WITHDRAWN",
"No. of patients with study drug withdrawn due to serious AE"
),
SERCONTRT = with_label(
AECONTRT == "Y" & AESER == "Y",
"No. of patients with dose modified/interrupted due to serious AE"
),
SERDSM = with_label(
AESER == "Y" & AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with treatment received for serious AE"
),
RELWD = with_label(
AEREL == "Y" & AEACN == "DRUG WITHDRAWN",
"No. of patients with study drug withdrawn due to related AE"
),
RELDSM = with_label(
AEREL == "Y" & AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with dose modified/interrupted due to related AE"
),
RELCONTRT = with_label(
AECONTRT == "Y" & AEREL == "Y",
"No. of patients with treatment received for related AE"
),
RELSER = with_label(
AESER == "Y" & AEREL == "Y",
"No. of patients with serious, related AE"
)
)
adae <- adae %>%
mutate(
AETOXGR = forcats::fct_recode(AETOXGR,
"Grade 1" = "1",
"Grade 2" = "2",
"Grade 3" = "3",
"Grade 4" = "4",
"Grade 5 (fatal outcome)" = "5"
)
)
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
::::::: panel-tabset
## Standard Table
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result")}
aesi_vars <- c("WD", "DSM", "CONTRT", "ALL_RESOLVED", "NOT_RESOLVED", "SER", "REL")
lyt_adae <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
count_patients_with_event(
vars = "USUBJID",
filters = c("ANL01FL" = "Y"),
denom = "N_col",
.labels = c(count_fraction = "Total number of patients with at least one AE")
) %>%
count_values(
"ANL01FL",
values = "Y",
.stats = "count",
.labels = c(count = "Total number of AEs"),
table_names = "total_aes"
) %>%
count_occurrences_by_grade(
var = "AETOXGR",
var_labels = "Total number of patients with at least one AE by worst grade",
show_labels = "visible"
) %>%
count_patients_with_flags("USUBJID", flag_variables = aesi_vars, denom = "N_col")
result <- build_table(lyt_adae, df = adae, alt_counts_df = adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with <br/> Optional Lines
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r variant2, test = list(result_v2 = "result")}
aesi_vars <- c("WD", "DSM", "CONTRT")
aesi_resolved <- c("ALL_RESOLVED", "ALL_RESOLVED_WD", "ALL_RESOLVED_DSM", "ALL_RESOLVED_CONTRT")
aesi_not_resolved <- c("NOT_RESOLVED", "NOT_RESOLVED_WD", "NOT_RESOLVED_DSM", "NOT_RESOLVED_CONTRT")
aesi_ser <- c("SER", "SERWD", "SERDSM", "SERCONTRT")
aesi_rel <- c("REL", "RELWD", "RELDSM", "RELCONTRT", "RELSER")
lyt_adae <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
count_patients_with_event(
vars = "USUBJID",
filters = c("ANL01FL" = "Y"),
denom = "N_col",
.labels = c(count_fraction = "Total number of patients with at least one AE")
) %>%
count_values(
"ANL01FL",
values = "Y",
.stats = "count",
.labels = c(count = "Total number of AEs"),
table_names = "total_aes"
) %>%
count_occurrences_by_grade(
var = "AETOXGR",
var_labels = "Total number of patients with at least one AE by worst grade",
show_labels = "visible"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = c(aesi_vars, aesi_resolved[1]), denom = "N_col"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_resolved[-1], denom = "N_col", .indent_mods = 1L, table_names = "fl_res"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_not_resolved[1], denom = "N_col", table_names = "fl_notres_main"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_not_resolved[-1], denom = "N_col", .indent_mods = 1L, table_names = "fl_notres"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_ser[1], denom = "N_col", table_names = "fl_ser_main"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_ser[-1], denom = "N_col", .indent_mods = 1L, table_names = "fl_ser"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_rel[1], denom = "N_col", table_names = "fl_rel_main"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_rel[-1], denom = "N_col", .indent_mods = 1L, table_names = "fl_rel"
)
result <- build_table(lyt_adae, df = adae, alt_counts_df = adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table For Studies <br/> with Multiple Drugs
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r variant3, test = list(result_v3 = "result")}
adsl <- random.cdisc.data::cadsl
adae_mult <- random.cdisc.data::cadae
adsl <- filter(adsl, SAFFL == "Y")
adae_mult <- filter(adae_mult, ANL01FL == "Y" & SAFFL == "Y")
adsl <- df_explicit_na(adsl)
adae_mult <- df_explicit_na(adae_mult)
# for illustration purposes only, create AEREL1, AEREL2, AEACN1, AEACN2 from respective variables
adae_mult <- adae_mult %>%
mutate(
AEREL1 = AEREL,
AEREL2 = AEREL,
AEACN1 = AEACN,
AEACN2 = AEACN
)
not_resolved <- adae_mult %>%
filter(!(AEOUT %in% c("RECOVERED/RESOLVED", "FATAL", "RECOVERED/RESOLVED WITH SEQUELAE"))) %>%
distinct(USUBJID) %>%
mutate(NOT_RESOLVED = "Y")
adae_mult <- adae_mult %>%
left_join(not_resolved, by = c("USUBJID")) %>%
mutate(
ALL_RESOLVED = with_label(
is.na(NOT_RESOLVED),
"Total number of patients with all non-fatal AEs resolved"
),
NOT_RESOLVED = with_label(
!is.na(NOT_RESOLVED),
"Total number of patients with at least one non-fatal unresolved or ongoing AE"
)
)
adae_mult <- adae_mult %>%
mutate(
AEDECOD = as.character(AEDECOD),
WD1 = with_label(
AEACN1 == "DRUG WITHDRAWN",
"Total number of patients with study drug 1 withdrawn due to AE"
),
WD2 = with_label(
AEACN2 == "DRUG WITHDRAWN",
"Total number of patients with study drug 2 withdrawn due to AE"
),
DSM1 = with_label(
AEACN1 %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"Total number of patients with dose of study drug 1 modified/interrupted due to AE"
),
DSM2 = with_label(
AEACN2 %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"Total number of patients with dose of study drug 2 modified/interrupted due to AE"
),
CONTRT = with_label(
AECONTRT == "Y",
"Total number of patients with treatment received for AE"
),
SER = with_label(
AESER == "Y",
"Total number of patients with at least one serious AE"
),
REL1 = with_label(
AEREL1 == "Y",
"Total number of patients with at least one AE related to study drug 1"
),
REL2 = with_label(
AEREL2 == "Y",
"Total number of patients with at least one AE related to study drug 2"
),
ALL_RESOLVED_WD1 = with_label(
WD1 == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with study drug 1 withdrawn due to resolved AE"
),
ALL_RESOLVED_DSM1 = with_label(
DSM1 == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with dose of study drug 1 modified/interrupted due to resolved AE"
),
ALL_RESOLVED_CONTRT = with_label(
CONTRT == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with treatment received for resolved AE"
),
ALL_RESOLVED_WD2 = with_label(
WD2 == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with study drug 2 withdrawn due to resolved AE"
),
ALL_RESOLVED_DSM2 = with_label(
DSM2 == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with dose of study drug 2 modified/interrupted due to resolved AE"
),
NOT_RESOLVED_WD1 = with_label(
WD1 == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with study drug 1 withdrawn due to unresolved or ongoing AE"
),
NOT_RESOLVED_DSM1 = with_label(
DSM1 == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with dose of study drug 1 modified/interrupted due to unresolved or ongoing AE"
),
NOT_RESOLVED_CONTRT = with_label(
CONTRT == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with treatment received for unresolved or ongoing AE"
),
NOT_RESOLVED_WD2 = with_label(
WD2 == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with study drug 2 withdrawn due to unresolved or ongoing AE"
),
NOT_RESOLVED_DSM2 = with_label(
DSM2 == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with dose of study drug 2 modified/interrupted due to unresolved or ongoing AE"
),
SERWD1 = with_label(
AESER == "Y" & AEACN1 == "DRUG WITHDRAWN",
"No. of patients with study drug 1 withdrawn due to serious AE"
),
SERWD2 = with_label(
AESER == "Y" & AEACN2 == "DRUG WITHDRAWN",
"No. of patients with study drug 2 withdrawn due to serious AE"
),
SERCONTRT = with_label(
AECONTRT == "Y" & AESER == "Y",
"No. of patients with treatment received for serious AE"
),
SERDSM1 = with_label(
AESER == "Y" & AEACN1 %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with dose of study drug 1 modified/interrupted due to serious AE"
),
SERDSM2 = with_label(
AESER == "Y" & AEACN2 %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with dose of study drug 2 modified/interrupted due to serious AE"
),
REL1WD1 = with_label(
AEREL1 == "Y" & AEACN1 == "DRUG WITHDRAWN",
"No. of patients with study drug 1 withdrawn due to AE related to study drug 1"
),
REL1WD2 = with_label(
AEREL1 == "Y" & AEACN2 == "DRUG WITHDRAWN",
"No. of patients with study drug 1 withdrawn due to AE related to study drug 2"
),
REL2WD1 = with_label(
AEREL1 == "Y" & AEACN1 == "DRUG WITHDRAWN",
"No. of patients with study drug 2 withdrawn due to AE related to study drug 1"
),
REL2WD2 = with_label(
AEREL1 == "Y" & AEACN2 == "DRUG WITHDRAWN",
"No. of patients with study drug 2 withdrawn due to AE related to study drug 2"
),
REL1DSM1 = with_label(
AEREL1 == "Y" & AEACN1 %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with dose of study drug 1 modified/interrupted due to AE related to study drug 1"
),
REL2DSM1 = with_label(
AEREL2 == "Y" & AEACN1 %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with dose of study drug 2 modified/interrupted due to AE related to study drug 1"
),
REL1DSM2 = with_label(
AEREL1 == "Y" & AEACN2 %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with dose of study drug 1 modified/interrupted due to AE related to study drug 2"
),
REL2DSM2 = with_label(
AEREL2 == "Y" & AEACN2 %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with dose of study drug 2 modified/interrupted due to AE related to study drug 2"
),
REL1CONTRT = with_label(
AECONTRT == "Y" & AEREL1 == "Y",
"No. of patients with treatment received for AE related to study drug 1"
),
REL2CONTRT = with_label(
AECONTRT == "Y" & AEREL2 == "Y",
"No. of patients with treatment received for AE related to study drug 2"
),
REL1SER = with_label(
AESER == "Y" & AEREL1 == "Y",
"No. of patients with serious AE related to study drug 1"
),
REL2SER = with_label(
AESER == "Y" & AEREL2 == "Y",
"No. of patients with serious AE related to study drug 2"
)
)
adae_mult <- adae_mult %>%
mutate(AETOXGR = forcats::fct_recode(
AETOXGR,
"Grade 1" = "1",
"Grade 2" = "2",
"Grade 3" = "3",
"Grade 4" = "4",
"Grade 5 (fatal outcome)" = "5"
))
aesi_vars <- c("WD1", "WD2", "DSM1", "DSM2", "CONTRT")
aesi_res <- c(
"ALL_RESOLVED",
"ALL_RESOLVED_WD1",
"ALL_RESOLVED_WD2",
"ALL_RESOLVED_DSM1",
"ALL_RESOLVED_DSM2",
"ALL_RESOLVED_CONTRT"
)
aesi_not_res <- c(
"NOT_RESOLVED",
"NOT_RESOLVED_WD1",
"NOT_RESOLVED_WD2",
"NOT_RESOLVED_DSM1",
"NOT_RESOLVED_DSM2",
"NOT_RESOLVED_CONTRT"
)
aesi_ser <- c("SER", "SERWD1", "SERWD2", "SERDSM1", "SERDSM2", "SERCONTRT")
aesi_rel1 <- c("REL1", "REL1WD1", "REL1WD2", "REL1DSM1", "REL1DSM2", "REL1CONTRT", "REL1SER")
aesi_rel2 <- c("REL2", "REL2WD1", "REL2WD2", "REL2DSM1", "REL2DSM2", "REL2CONTRT", "REL2SER")
lyt_adae_mult <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
count_patients_with_event(
vars = "USUBJID",
filters = c("ANL01FL" = "Y"),
denom = "N_col",
.labels = c(count_fraction = "Total number of patients with at least one AE")
) %>%
count_values(
"ANL01FL",
values = "Y",
.stats = "count",
.labels = c(count = "Total number of AEs"),
table_names = "total_aes"
) %>%
count_occurrences_by_grade(
var = "AETOXGR",
var_labels = "Total number of patients with at least one AE by worst grade",
show_labels = "visible"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = c(aesi_vars, aesi_res[1]), denom = "N_col"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_res[-1], denom = "N_col", .indent_mods = 1L, table_names = "fl_res"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_not_res[1], denom = "N_col", table_names = "fl_notres_main"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_not_res[-1], denom = "N_col", .indent_mods = 1L, table_names = "fl_notres"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_ser[1], denom = "N_col", table_names = "fl_ser_main"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_ser[-1], denom = "N_col", .indent_mods = 1L, table_names = "fl_ser"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_rel1[1], denom = "N_col", table_names = "fl_rel1_main"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_rel1[-1], denom = "N_col", .indent_mods = 1L, table_names = "fl_rel1"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_rel2[1], denom = "N_col", table_names = "fl_rel2_main"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_rel2[-1], denom = "N_col", .indent_mods = 1L, table_names = "fl_rel2"
)
result <- build_table(lyt_adae_mult, df = adae_mult, alt_counts_df = adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant3")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table of AEs <br/> by SMQ
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r variant4, test = list(result_v4 = "result")}
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
adsl <- filter(adsl, SAFFL == "Y")
adae <- filter(adae, ANL01FL == "Y" & SAFFL == "Y")
adsl <- df_explicit_na(adsl)
adae <- df_explicit_na(adae)
stack_adae_by_smq <- function(adae, smq) {
adae_labels <- c(var_labels(adae), "Standardized MedDRA Query")
l_df <- lapply(smq, function(ae_grp) {
ae_scope <- gsub("NAM", "SC", ae_grp)
keep <- adae[[ae_grp]] != "<Missing>"
df <- adae[keep, ]
if (substr(ae_grp, 1, 3) == "SMQ") {
df[["SMQ"]] <- aesi_label(as.character(df[[ae_grp]]), scope = as.character(df[[ae_scope]]))
} else {
df[["SMQ"]] <- df[[ae_grp]]
}
df
})
result <- do.call(rbind, l_df)
var_labels(result) <- adae_labels
result
}
adae_smq <- stack_adae_by_smq(adae, c("SMQ01NAM", "SMQ02NAM", "CQ01NAM"))
not_resolved <- adae_smq %>%
filter(!(AEOUT %in% c("RECOVERED/RESOLVED", "FATAL", "RECOVERED/RESOLVED WITH SEQUELAE"))) %>%
distinct(USUBJID) %>%
mutate(NOT_RESOLVED = "Y")
adae_smq <- adae_smq %>%
left_join(not_resolved, by = c("USUBJID")) %>%
mutate(
ALL_RESOLVED = with_label(
is.na(NOT_RESOLVED),
"Total number of patients with all non-fatal AEs resolved"
),
NOT_RESOLVED = with_label(
!is.na(NOT_RESOLVED),
"Total number of patients with at least one non-fatal unresolved or ongoing AE"
)
)
adae_smq <- adae_smq %>%
mutate(
AEDECOD = as.character(AEDECOD),
WD = with_label(
AEACN == "DRUG WITHDRAWN",
"Total number of patients with study drug withdrawn due to AE"
),
DSM = with_label(
AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"Total number of patients with dose modified/interrupted due to AE"
),
CONTRT = with_label(
AECONTRT == "Y",
"Total number of patients with treatment received for AE"
),
SER = with_label(
AESER == "Y",
"Total number of patients with at least one serious AE"
),
REL = with_label(
AEREL == "Y",
"Total number of patients with at least one related AE"
),
ALL_RESOLVED_WD = with_label(
WD == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with study drug withdrawn due to resolved AE"
),
ALL_RESOLVED_DSM = with_label(
DSM == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with dose modified/interrupted due to resolved AE"
),
ALL_RESOLVED_CONTRT = with_label(
CONTRT == TRUE & ALL_RESOLVED == TRUE,
"No. of patients with treatment received for resolved AE"
),
NOT_RESOLVED_WD = with_label(
WD == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with study drug withdrawn due to unresolved or ongoing AE"
),
NOT_RESOLVED_DSM = with_label(
DSM == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with dose modified/interrupted due to unresolved or ongoing AE"
),
NOT_RESOLVED_CONTRT = with_label(
CONTRT == TRUE & NOT_RESOLVED == TRUE,
"No. of patients with treatment received for unresolved or ongoing AE"
),
SERWD = with_label(
AESER == "Y" & AEACN == "DRUG WITHDRAWN",
"No. of patients with study drug withdrawn due to serious AE"
),
SERCONTRT = with_label(
AECONTRT == "Y" & AESER == "Y",
"No. of patients with treatment received for serious AE"
),
SERDSM = with_label(
AESER == "Y" & AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with dose modified/interrupted due to serious AE"
),
RELWD = with_label(
AEREL == "Y" & AEACN == "DRUG WITHDRAWN",
"No. of patients with study drug withdrawn due to related AE"
),
RELDSM = with_label(
AEREL == "Y" & AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"No. of patients with dose modified/interrupted due to related AE"
),
RELCONTRT = with_label(
AECONTRT == "Y" & AEREL == "Y",
"No. of patients with treatment received for related AE"
),
RELSER = with_label(
AESER == "Y" & AEREL == "Y",
"No. of patients with serious, related AE"
)
)
adae_smq <- adae_smq %>%
mutate(
AETOXGR = forcats::fct_recode(AETOXGR,
"Grade 1" = "1",
"Grade 2" = "2",
"Grade 3" = "3",
"Grade 4" = "4",
"Grade 5 (fatal outcome)" = "5"
)
)
split_fun <- remove_split_levels("<Missing>")
aesi_vars <- c("WD", "DSM", "CONTRT", "ALL_RESOLVED", "NOT_RESOLVED", "SER", "REL")
lyt_adae <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
split_rows_by(
"SMQ",
child_labels = "visible",
split_fun = split_fun,
split_label = "Standardized MedDRA Query",
label_pos = "topleft"
) %>%
count_patients_with_event(
vars = "USUBJID",
filters = c("ANL01FL" = "Y"),
denom = "N_col",
.labels = c(count_fraction = "Total number of patients with at least one AE")
) %>%
count_values(
"ANL01FL",
values = "Y",
.stats = "count",
.labels = c(count = "Total number of AEs"),
table_names = "total_aes"
) %>%
count_occurrences_by_grade(
var = "AETOXGR",
var_labels = "Total number of patients with at least one AE by worst grade",
.show_labels = "visible"
) %>%
count_patients_with_flags("USUBJID", flag_variables = aesi_vars, denom = "N_col")
result <- build_table(lyt_adae, df = adae_smq, alt_counts_df = adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant4")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
To illustrate, additional variables such as flags (TRUE/FALSE) for selected AEs of interest. Please consult your SAP on how to handle missing AE grades.
```{r setup}
#| code-fold: show
```
:::::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
library(dplyr)
ADSL <- random.cdisc.data::cadsl
ADAE <- random.cdisc.data::cadae
ADAE <- filter(ADAE, ANL01FL == "Y" & SAFFL == "Y")
not_resolved <- ADAE %>%
filter(!(AEOUT %in% c("RECOVERED/RESOLVED", "FATAL", "RECOVERED/RESOLVED WITH SEQUELAE"))) %>%
distinct(USUBJID) %>%
mutate(NOT_RESOLVED = "Y")
ADAE <- ADAE %>%
left_join(not_resolved, by = c("USUBJID")) %>%
mutate(
ALL_RESOLVED = with_label(is.na(NOT_RESOLVED), "All non-fatal AEs resolved"),
NOT_RESOLVED = with_label(!is.na(NOT_RESOLVED), "At least one non-fatal unresolved or ongoing AE")
)
ADAE <- ADAE %>%
mutate(
AEDECOD = as.character(AEDECOD),
WD = with_label(AEACN == "DRUG WITHDRAWN", "AE that led to study drug withdrawal"),
DSM = with_label(
AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"AE that led to study drug dose modified/interrupted"
),
CONTRT = with_label(AECONTRT == "Y", "AE that required treatment"),
SER = with_label(AESER == "Y", "Serious AE"),
REL = with_label(AEREL == "Y", "Related AE"),
ALL_RESOLVED_WD = with_label(
WD == TRUE & ALL_RESOLVED == TRUE,
"Resolved AE that led to study drug withdrawal"
),
ALL_RESOLVED_DSM = with_label(
DSM == TRUE & ALL_RESOLVED == TRUE,
"Resolved AE that led to study drug dose modified/interrupted"
),
ALL_RESOLVED_CONTRT = with_label(
CONTRT == TRUE & ALL_RESOLVED == TRUE,
"Resolved AE that required treatment"
),
NOT_RESOLVED_WD = with_label(
WD == TRUE & NOT_RESOLVED == TRUE,
"Unresolved AE that led to study drug withdrawal"
),
NOT_RESOLVED_DSM = with_label(
DSM == TRUE & NOT_RESOLVED == TRUE,
"Unresolved AE that led to study drug dose modified/interrupted"
),
NOT_RESOLVED_CONTRT = with_label(
CONTRT == TRUE & NOT_RESOLVED == TRUE,
"Unresolved AE that required treatment"
),
SERWD = with_label(
AESER == "Y" & AEACN == "DRUG WITHDRAWN",
"Serious AE that led to study drug withdrawal"
),
SERCONTRT = with_label(
AECONTRT == "Y" & AESER == "Y",
"Serious AE that required treatment"
),
SERDSM = with_label(
AESER == "Y" & AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"Serious AE that led to study drug dose modified/interrupted"
),
RELWD = with_label(
AEREL == "Y" & AEACN == "DRUG WITHDRAWN", "Related AE that led to study drug withdrawal"
),
RELDSM = with_label(
AEREL == "Y" & AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"Related AE that led to study drug dose modified/interrupted"
),
RELCONTRT = with_label(AECONTRT == "Y" & AEREL == "Y", "Related AE that required treatment"),
RELSER = with_label(AESER == "Y" & AEREL == "Y", "Serious related AE")
)
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADAE")]
aesi_vars <- c(
"WD", "DSM", "CONTRT", "ALL_RESOLVED_WD", "ALL_RESOLVED_DSM", "ALL_RESOLVED_CONTRT",
"NOT_RESOLVED_WD", "NOT_RESOLVED_DSM", "NOT_RESOLVED_CONTRT", "SER", "SERWD", "SERDSM",
"SERCONTRT", "REL", "RELWD", "RELDSM", "RELCONTRT", "RELSER"
)
## Setup App
app <- init(
data = data,
modules = modules(
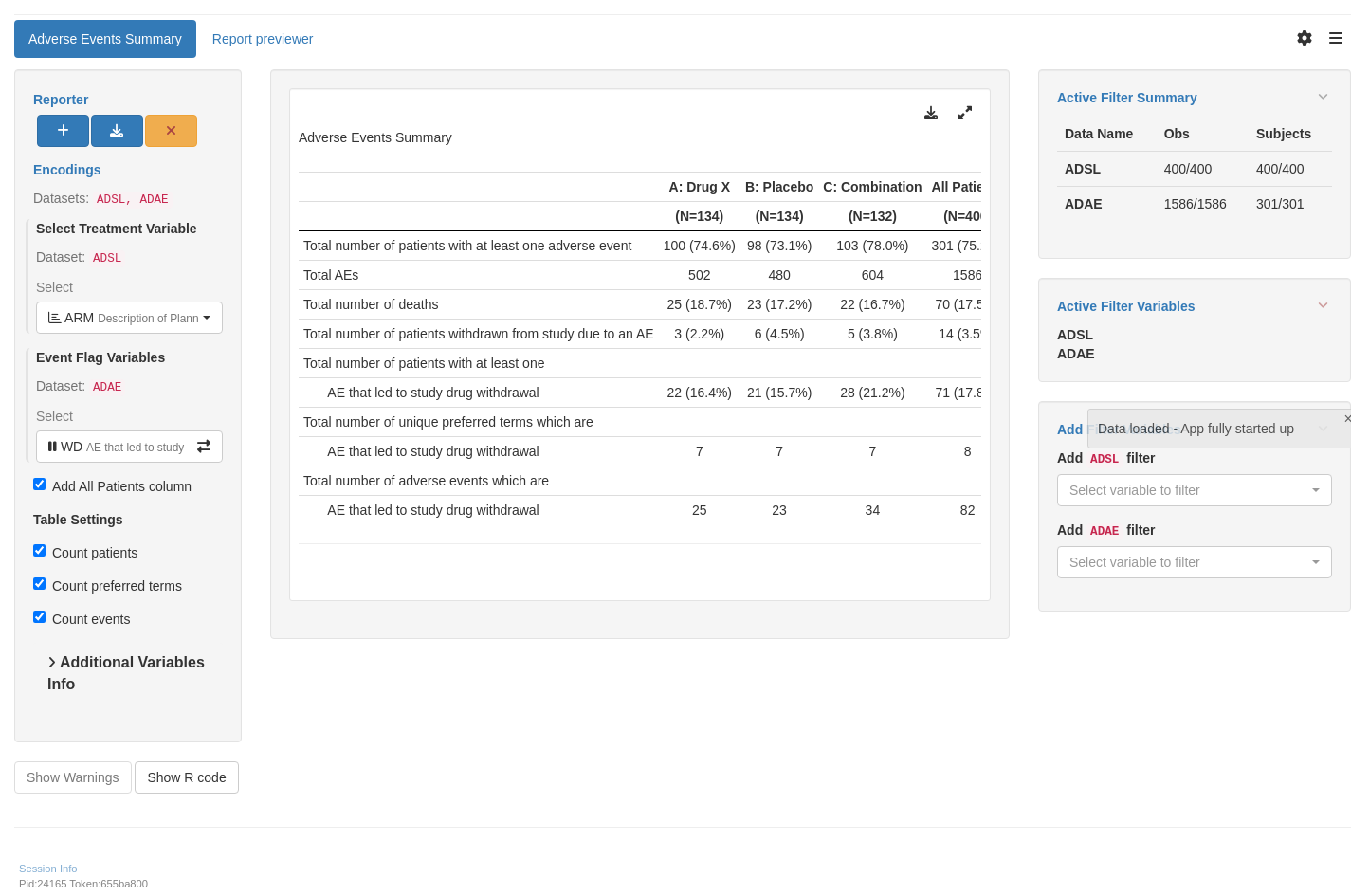
tm_t_events_summary(
label = "Adverse Events Summary",
dataname = "ADAE",
arm_var = choices_selected(
choices = variable_choices("ADSL", c("ARM", "ARMCD")),
selected = "ARM"
),
flag_var_anl = choices_selected(
choices = variable_choices("ADAE", aesi_vars),
selected = aesi_vars[1],
keep_order = TRUE,
fixed = FALSE
),
add_total = TRUE
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}