---
title: DST01
subtitle: Patient Disposition
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(tern)
library(dplyr)
set.seed(1, kind = "Mersenne-Twister")
adsl <- random.cdisc.data::cadsl
# reorder EOSSTT factor levels so DISCONTINUED is the last level
adsl <- df_explicit_na(adsl) %>%
mutate(EOSSTT = factor(EOSSTT, levels = c("COMPLETED", "ONGOING", "DISCONTINUED")))
adsl_gp_added <- adsl %>%
mutate(DCSREASGP = case_when(
DCSREAS %in% c("ADVERSE EVENT", "DEATH") ~ "Safety",
(DCSREAS != "<Missing>" & !DCSREAS %in% c("ADVERSE EVENT", "DEATH")) ~ "Non-Safety",
DCSREAS == "<Missing>" ~ "<Missing>"
) %>% factor(levels = c("Safety", "Non-Safety", "<Missing>")))
adsl_eotstt_added <- adsl_gp_added %>%
mutate(
EOTSTT = sample(
c("ONGOING", "COMPLETED", "DISCONTINUED"),
size = nrow(adsl),
replace = TRUE
) %>% factor(levels = c("COMPLETED", "ONGOING", "DISCONTINUED"))
)
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
:::::: panel-tabset
## Standard Table
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result1")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(
"ACTARM",
split_fun = add_overall_level("All Patients", first = FALSE)
) %>%
count_occurrences(
"EOSSTT",
show_labels = "hidden"
) %>%
analyze_vars(
"DCSREAS",
.stats = "count_fraction",
denom = "N_col",
show_labels = "hidden",
.indent_mods = c(count_fraction = 1L)
)
result1 <- build_table(lyt = lyt, df = adsl)
result1
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Grouping of Reasons
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant2, test = list(result_v2 = "result2")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(
"ACTARM",
split_fun = add_overall_level("All Patients", first = FALSE)
) %>%
count_occurrences(
"EOSSTT",
show_labels = "hidden"
) %>%
split_rows_by("DCSREASGP", indent_mod = 1L) %>%
analyze_vars(
"DCSREAS",
.stats = "count_fraction",
denom = "N_col",
show_labels = "hidden"
)
tbl <- build_table(lyt = lyt, df = adsl_gp_added)
result2 <- prune_table(tbl) # remove rows containing all zeros
result2
```
```{r include = FALSE}
webr_code_labels <- c("variant2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table Adding Optional Rows
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant3, test = list(result_v3 = "result3")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(
"ACTARM",
split_fun = add_overall_level("All Patients", first = FALSE)
) %>%
count_occurrences(
"EOTSTT",
show_labels = "hidden"
)
tbl <- build_table(lyt = lyt, df = adsl_eotstt_added)
tbl <- prune_table(tbl) # remove rows containing all zeros
# Combine tables
col_info(result2) <- col_info(tbl)
result3 <- rbind(result2, tbl)
result3
```
```{r include = FALSE}
webr_code_labels <- c("variant3")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
::::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
library(dplyr)
set.seed(1, kind = "Mersenne-Twister")
ADSL <- random.cdisc.data::cadsl
ADSL <- df_explicit_na(ADSL)
ADSL <- ADSL %>%
mutate(
DCSREASGP = case_when(
DCSREAS %in% c("ADVERSE EVENT", "DEATH") ~ "Safety",
(DCSREAS != "<Missing>" & !DCSREAS %in% c("ADVERSE EVENT", "DEATH")) ~ "Non-Safety",
DCSREAS == "<Missing>" ~ "<Missing>"
) %>% as.factor(),
EOTSTT = sample(
c("ONGOING", "COMPLETED", "DISCONTINUED"),
size = nrow(ADSL),
replace = TRUE
) %>% as.factor()
) %>%
col_relabel(
EOTSTT = "End Of Treatment Status"
)
date_vars_asl <- names(ADSL)[vapply(ADSL, function(x) inherits(x, c("Date", "POSIXct", "POSIXlt")), logical(1))]
demog_vars_asl <- names(ADSL)[!(names(ADSL) %in% c("USUBJID", "STUDYID", date_vars_asl))]
})
join_keys(data) <- default_cdisc_join_keys["ADSL"]
## Reusable Configuration For Modules
ADSL <- data[["ADSL"]]
demog_vars_asl <- data[["demog_vars_asl"]]
## Setup App
app <- init(
data = data,
modules = modules(
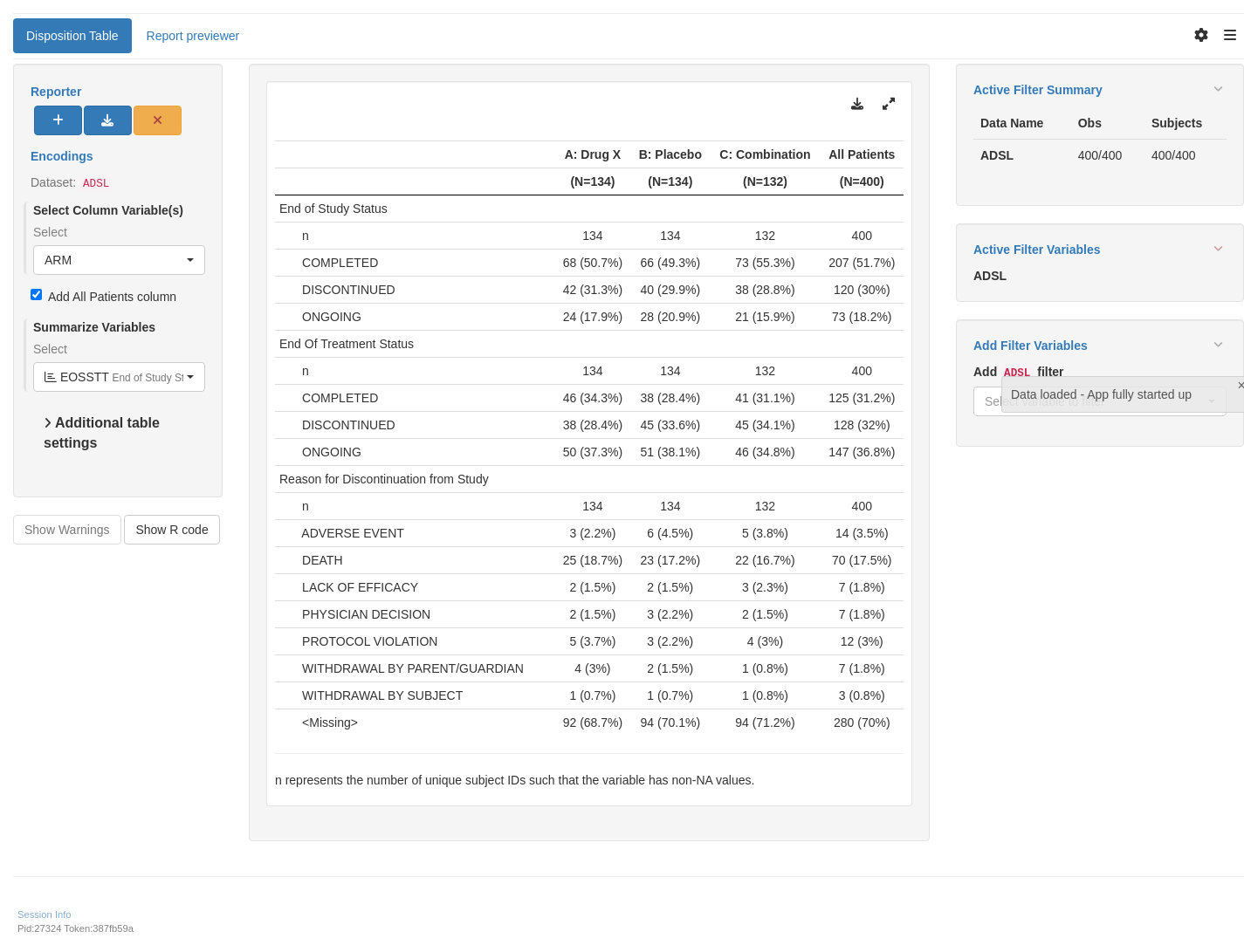
tm_t_summary(
label = "Disposition Table",
dataname = "ADSL",
arm_var = choices_selected(c("ARM", "ARMCD"), "ARM"),
summarize_vars = choices_selected(
variable_choices(ADSL, demog_vars_asl),
c("EOSSTT", "DCSREAS", "EOTSTT")
),
useNA = "ifany"
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}