RNAG9
RNAseq Kaplan-Meier Graph
This page can be used as a template of how to produce Kaplan-Meier graphs for RNA-seq gene expression analysis using available tern and hermes functions, and to create an interactive Kaplan-Meier graph for RNA-seq gene expression analysis using teal.modules.hermes.
The code below needs both RNA-seq data (in HermesData format) and time-to-event data (in ADTTE format) as input.
We first prepare the time-to-event data. We define an event indicator variable, transform the time to months and filter down to the overall survival subset.
Then we prepare the RNA-seq data. See RNAG1 for basic details on how to import, filter and normalize HermesData. We use col_data_with_genes() to extract the sample variables (colData) from the object, together with a single specified gene or a specified gene signature. See ?hermes::gene_spec for details on how to do this. Then we use inner_join_cdisc() to join this genetic data with the ADTTE data from above. See the help page for more details, in particular how the join keys can be customized if needed - here we just join based on USUBJID by default.
We can then cut the resulting gene column (we figure out the column name and save it in arm_name below) in the joined_data into quantile bins (in this example we want three equally sized groups).
It is now simple to create the Kaplan-Meier graph by providing the data set created above with the variable specification. Note that we specify the above created gene_factor as arm variable here.
Code
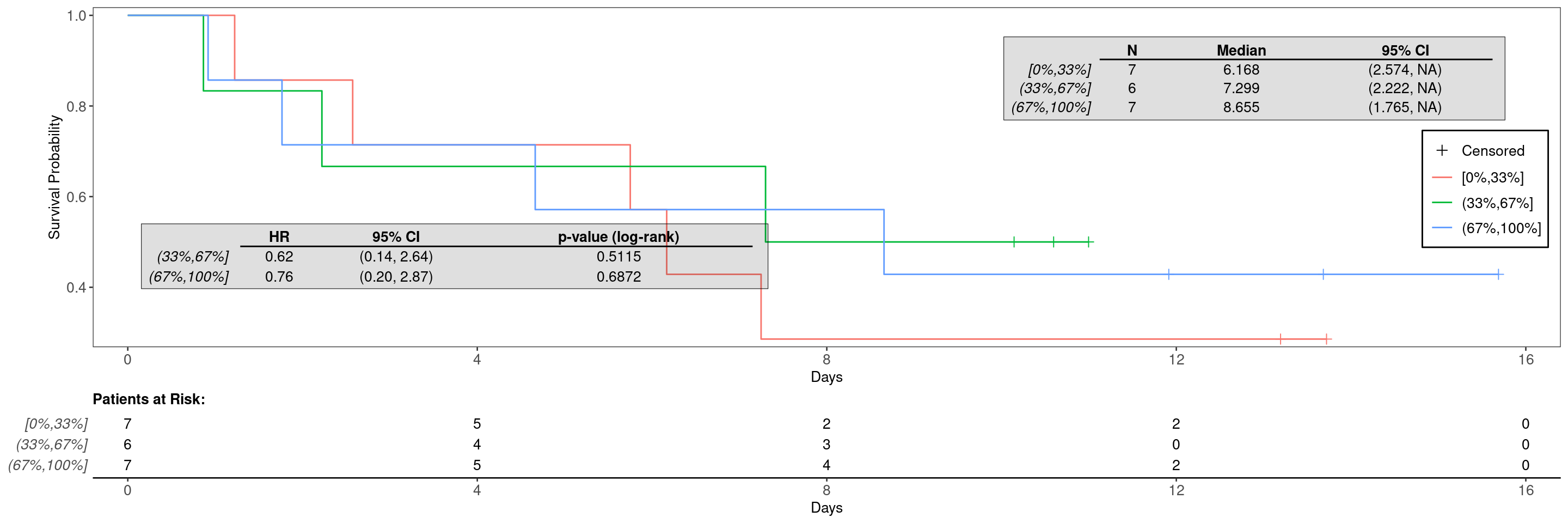
See KG1 to KG5 for additional customization options for the Kaplan-Meier graphs or the help page ?g_km().
We start by importing a MultiAssayExperiment and sample ADTTE data; here we use the example multi_assay_experiment available in hermes and example ADTTE data from random.cdisc.data. We can then use the provided teal module tm_g_km to include the corresponding interactive Kaplan-Meier analysis in our teal app. Note that by default the counts assay is excluded via the exclude_assays argument, but we can include it by just saying that we don’t want to exclude any assays. In case that we have different non-standard column names in our ADTTE data set we could also specify them via the adtte_vars argument, see the documentation ?tm_g_km for more details.
Code
Warning: `datanames<-()` was deprecated in teal.data 0.7.0.
ℹ invalid to use `datanames()<-` or `names()<-` on an object of class
`teal_data`. See ?names.teal_dataCode
[INFO] 2025-02-19 17:26:28.7126 pid:3180 token:[] teal.modules.hermes Initializing tm_g_kmWarning: 'experiments' dropped; see 'drops()'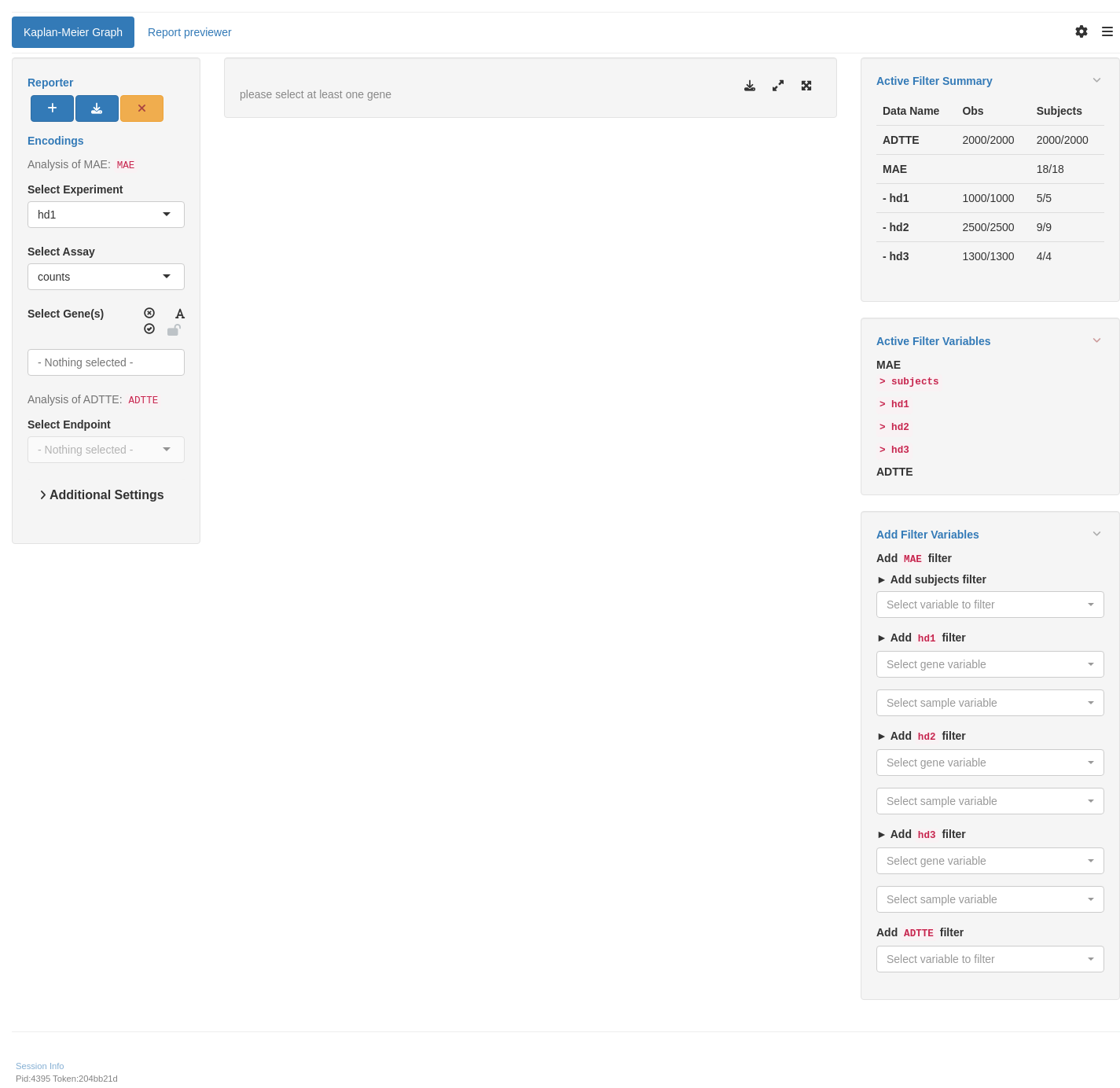
R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] random.cdisc.data_0.3.16 teal.modules.hermes_0.1.6
[3] teal_0.16.0 teal.slice_0.6.0
[5] teal.data_0.7.0 teal.code_0.6.1
[7] shiny_1.10.0 hermes_1.10.0
[9] SummarizedExperiment_1.36.0 Biobase_2.66.0
[11] GenomicRanges_1.58.0 GenomeInfoDb_1.42.3
[13] IRanges_2.40.1 S4Vectors_0.44.0
[15] BiocGenerics_0.52.0 MatrixGenerics_1.18.1
[17] matrixStats_1.5.0 ggfortify_0.4.17
[19] ggplot2_3.5.1 dplyr_1.1.4
[21] tern_0.9.7 rtables_0.6.11
[23] magrittr_2.0.3 formatters_0.5.10
loaded via a namespace (and not attached):
[1] RColorBrewer_1.1-3 jsonlite_1.9.0
[3] shape_1.4.6.1 nestcolor_0.1.3
[5] MultiAssayExperiment_1.32.0 farver_2.1.2
[7] rmarkdown_2.29 GlobalOptions_0.1.2
[9] zlibbioc_1.52.0 vctrs_0.6.5
[11] memoise_2.0.1 webshot_0.5.5
[13] BiocBaseUtils_1.9.0 htmltools_0.5.8.1
[15] S4Arrays_1.6.0 forcats_1.0.0
[17] progress_1.2.3 curl_6.2.1
[19] broom_1.0.7 SparseArray_1.6.1
[21] sass_0.4.9 bslib_0.9.0
[23] fontawesome_0.5.3 htmlwidgets_1.6.4
[25] httr2_1.1.0 cachem_1.1.0
[27] teal.widgets_0.4.3 mime_0.12
[29] lifecycle_1.0.4 iterators_1.0.14
[31] pkgconfig_2.0.3 webshot2_0.1.1
[33] Matrix_1.7-2 R6_2.6.1
[35] fastmap_1.2.0 GenomeInfoDbData_1.2.13
[37] rbibutils_2.3 clue_0.3-66
[39] digest_0.6.37 colorspace_2.1-1
[41] shinycssloaders_1.1.0 ps_1.9.0
[43] AnnotationDbi_1.68.0 RSQLite_2.3.9
[45] filelock_1.0.3 labeling_0.4.3
[47] httr_1.4.7 abind_1.4-8
[49] compiler_4.4.2 bit64_4.6.0-1
[51] withr_3.0.2 doParallel_1.0.17
[53] backports_1.5.0 DBI_1.2.3
[55] logger_0.4.0 biomaRt_2.62.1
[57] rappdirs_0.3.3 DelayedArray_0.32.0
[59] rjson_0.2.23 chromote_0.4.0
[61] tools_4.4.2 httpuv_1.6.15
[63] glue_1.8.0 callr_3.7.6
[65] promises_1.3.2 grid_4.4.2
[67] checkmate_2.3.2 cluster_2.1.8
[69] generics_0.1.3 gtable_0.3.6
[71] websocket_1.4.2 tidyr_1.3.1
[73] hms_1.1.3 xml2_1.3.6
[75] XVector_0.46.0 ggrepel_0.9.6
[77] foreach_1.5.2 pillar_1.10.1
[79] stringr_1.5.1 later_1.4.1
[81] circlize_0.4.16 splines_4.4.2
[83] BiocFileCache_2.14.0 lattice_0.22-6
[85] survival_3.8-3 bit_4.5.0.1
[87] tidyselect_1.2.1 ComplexHeatmap_2.22.0
[89] Biostrings_2.74.1 knitr_1.49
[91] gridExtra_2.3 teal.logger_0.3.2
[93] xfun_0.51 stringi_1.8.4
[95] UCSC.utils_1.2.0 yaml_2.3.10
[97] shinyWidgets_0.8.7 evaluate_1.0.3
[99] codetools_0.2-20 tibble_3.2.1
[101] cli_3.6.4 xtable_1.8-4
[103] Rdpack_2.6.2 jquerylib_0.1.4
[105] processx_3.8.5 munsell_0.5.1
[107] teal.reporter_0.4.0 Rcpp_1.0.14
[109] dbplyr_2.5.0 png_0.1-8
[111] parallel_4.4.2 assertthat_0.2.1
[113] blob_1.2.4 prettyunits_1.2.0
[115] scales_1.3.0 purrr_1.0.4
[117] crayon_1.5.3 GetoptLong_1.0.5
[119] rlang_1.1.5 formatR_1.14
[121] cowplot_1.1.3 KEGGREST_1.46.0
[123] shinyjs_2.1.0