DG3A
Barplot of a Categorical Variable by Another Categorical Variable
The graphs below summarize the distribution of a categorical biomarker variable as barplots, either in the overall population or by one or more categorical clinical variables.
We will use the cadsl data set from the random.cdisc.data package to illustrate the graph and will select on the biomarker evaluable population with BEP01FL. The column BMRKR2 contains the biomarker values on a categorical scale. We will use ARM as categorical clinical variable.
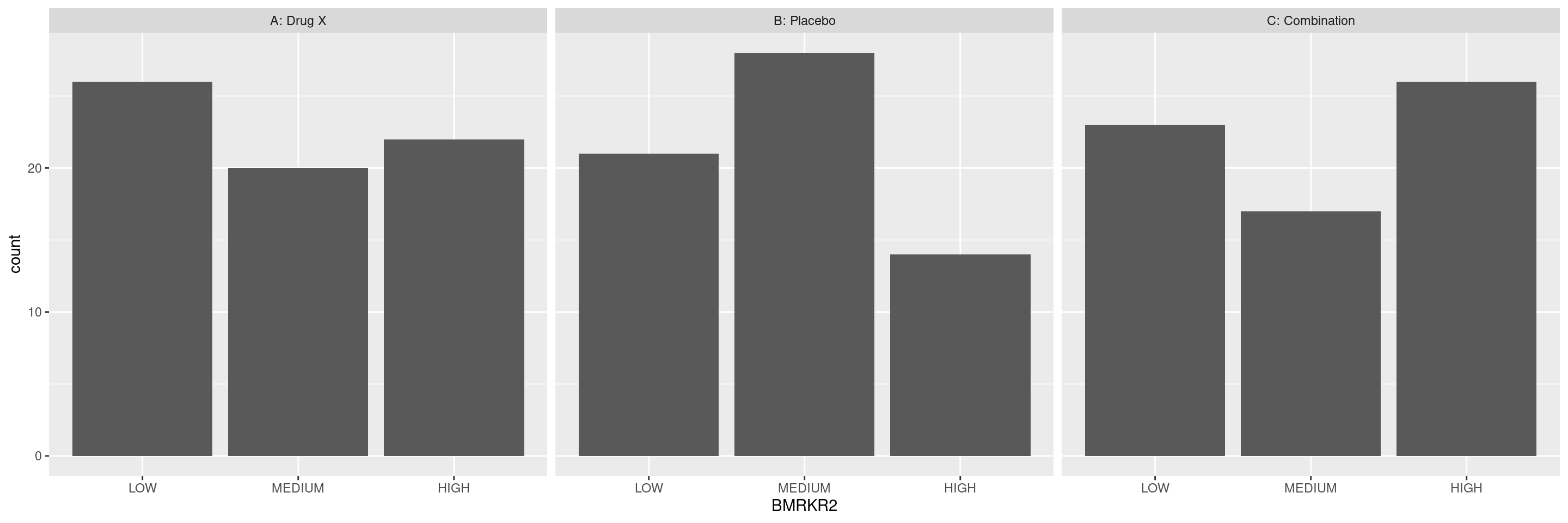
Here below the code for a simple distribution of the category counts of a first biomarker variable (BMRKR2) split by a second categorical variable (ARM). We can use again the facet_grid() layer.
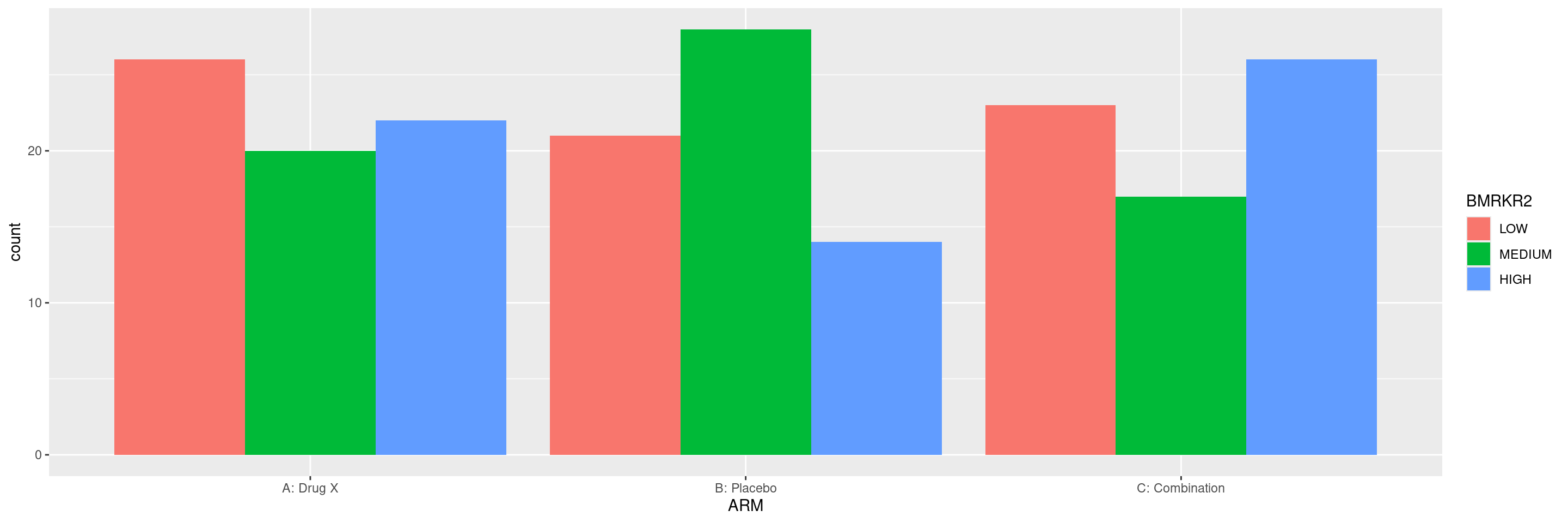
We could instead display the columns next to each other within the same graph with the fill aesthetic and the position_dodge() option instead of using the facet_grid() layer.
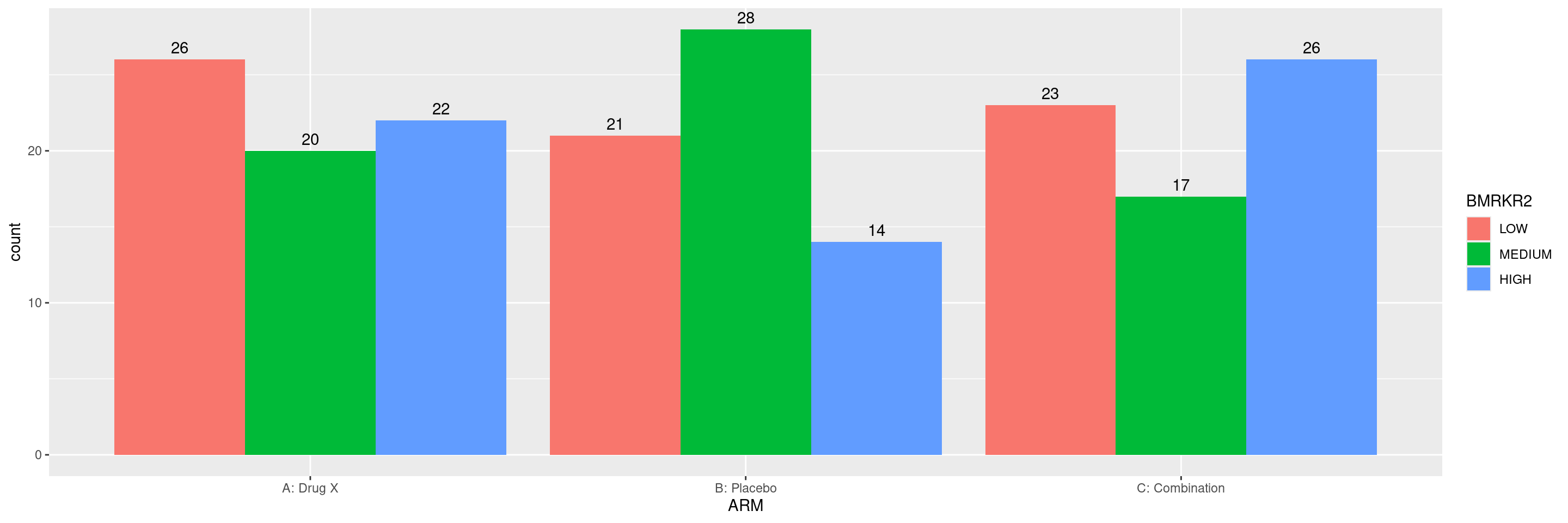
We can then again add the absolute count above each of the columns.
R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_1.1.4 ggplot2.utils_0.3.2 ggplot2_3.5.1
[4] tern_0.9.7 rtables_0.6.11 magrittr_2.0.3
[7] formatters_0.5.10
loaded via a namespace (and not attached):
[1] generics_0.1.3 tidyr_1.3.1 EnvStats_3.0.0
[4] stringi_1.8.4 lattice_0.22-6 digest_0.6.37
[7] evaluate_1.0.3 grid_4.4.2 fastmap_1.2.0
[10] jsonlite_1.9.0 Matrix_1.7-2 backports_1.5.0
[13] survival_3.8-3 purrr_1.0.4 scales_1.3.0
[16] codetools_0.2-20 Rdpack_2.6.2 cli_3.6.4
[19] ggpp_0.5.8-1 nestcolor_0.1.3 rlang_1.1.5
[22] rbibutils_2.3 munsell_0.5.1 splines_4.4.2
[25] withr_3.0.2 yaml_2.3.10 tools_4.4.2
[28] polynom_1.4-1 checkmate_2.3.2 colorspace_2.1-1
[31] forcats_1.0.0 ggstats_0.8.0 broom_1.0.7
[34] vctrs_0.6.5 R6_2.6.1 lifecycle_1.0.4
[37] stringr_1.5.1 htmlwidgets_1.6.4 MASS_7.3-64
[40] pkgconfig_2.0.3 pillar_1.10.1 gtable_0.3.6
[43] glue_1.8.0 xfun_0.51 tibble_3.2.1
[46] tidyselect_1.2.1 knitr_1.49 farver_2.1.2
[49] htmltools_0.5.8.1 labeling_0.4.3 rmarkdown_2.29
[52] random.cdisc.data_0.3.16 compiler_4.4.2