RG1B
Change Color Scheme of Response Graph for Overall Population
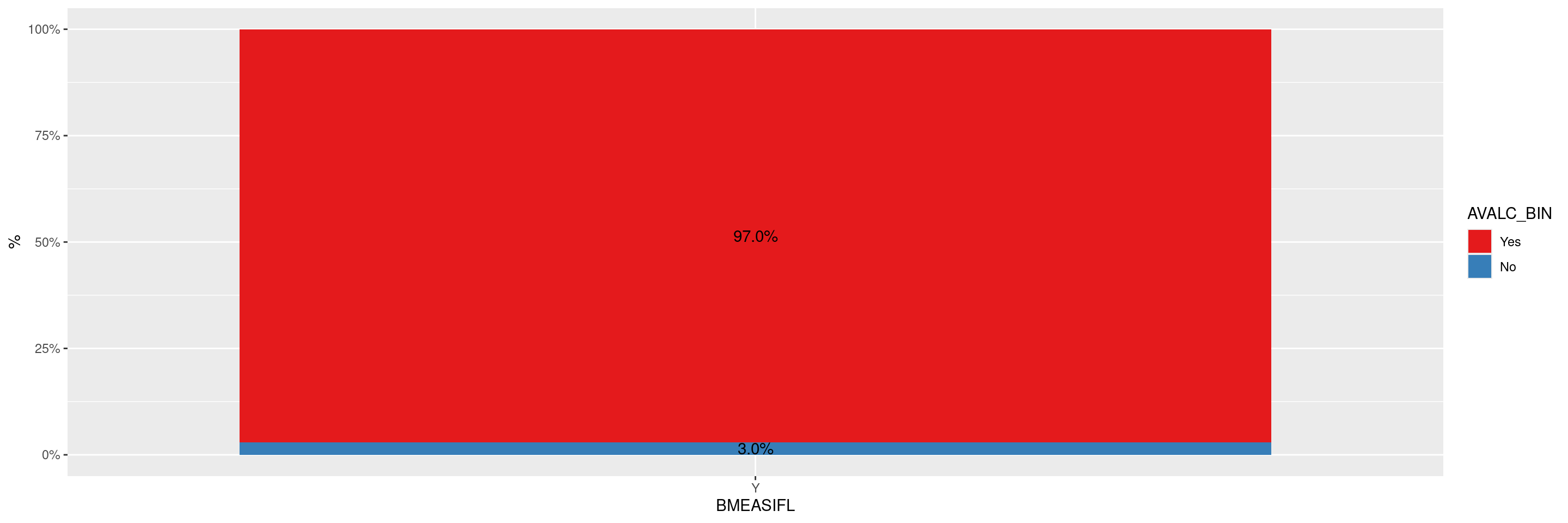
We will use the cadrs data set from the random.cdisc.data package to create the response plots. We transform the response variable into an ordered factor to ensure that the response labels are ordered correctly and a sequential color scheme is used in the graph. We select Best Confirmed Overall Response by Investigator to evaluate response. Finally, we select patients with measurable disease at baseline (BMEASIFL == "Y") as response evaluable patients.
For ggplot() used in all analyses, we add by = BMEASIFL in the aesthetics to support the calculation of proportions using geom_text(stat = "prop").
We can use the scale_fill_manual() function from ggplot2 to change the color scheme of the plot by manually selecting colors to assign to the values.
Code
adrs <- adrs %>%
mutate(
AVALC_BIN = fct_collapse_only(
AVALC,
Yes = c("CR", "PR"),
No = c("PD", "SD", "NE", "<Missing>")
)
)
graph <- ggplot(adrs, aes(BMEASIFL, fill = AVALC_BIN, by = BMEASIFL)) +
geom_bar(position = "fill") +
geom_text(stat = "prop", position = position_fill(.5)) +
scale_y_continuous(labels = scales::percent) +
ylab("%")
graph +
scale_fill_manual("AVALC_BIN", values = c("Yes" = "blue", "No" = "red"))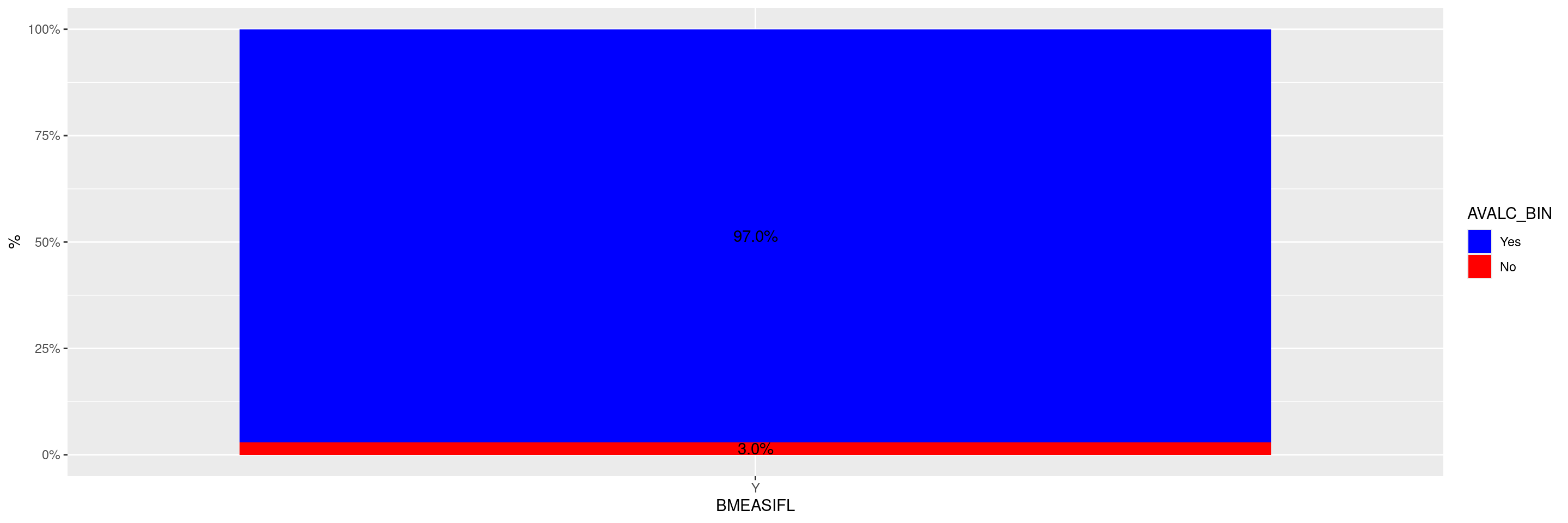
Alternatively we can also use the scale_fill_brewer() function from ggplot2 to change the color scheme of the plot by selecting a preset color palette.
R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_1.1.4 ggplot2.utils_0.3.2 ggplot2_3.5.1
[4] tern_0.9.7 rtables_0.6.11 magrittr_2.0.3
[7] formatters_0.5.10
loaded via a namespace (and not attached):
[1] generics_0.1.3 tidyr_1.3.1 EnvStats_3.0.0
[4] stringi_1.8.4 lattice_0.22-6 digest_0.6.37
[7] RColorBrewer_1.1-3 evaluate_1.0.3 grid_4.4.2
[10] fastmap_1.2.0 jsonlite_1.9.0 Matrix_1.7-2
[13] backports_1.5.0 survival_3.8-3 purrr_1.0.4
[16] scales_1.3.0 codetools_0.2-20 Rdpack_2.6.2
[19] cli_3.6.4 ggpp_0.5.8-1 nestcolor_0.1.3
[22] rlang_1.1.5 rbibutils_2.3 munsell_0.5.1
[25] splines_4.4.2 withr_3.0.2 yaml_2.3.10
[28] tools_4.4.2 polynom_1.4-1 checkmate_2.3.2
[31] colorspace_2.1-1 forcats_1.0.0 ggstats_0.8.0
[34] broom_1.0.7 vctrs_0.6.5 R6_2.6.1
[37] lifecycle_1.0.4 stringr_1.5.1 htmlwidgets_1.6.4
[40] MASS_7.3-64 pkgconfig_2.0.3 pillar_1.10.1
[43] gtable_0.3.6 glue_1.8.0 xfun_0.51
[46] tibble_3.2.1 tidyselect_1.2.1 knitr_1.49
[49] farver_2.1.2 htmltools_0.5.8.1 labeling_0.4.3
[52] rmarkdown_2.29 random.cdisc.data_0.3.16 compiler_4.4.2