SFG3A
Comparing Between Genders in Survival Forest Graph for One Treatment Arm
SFG
We prepare the data similarly as in SFG1, focusing on a single arm in the biomarker evaluable population.
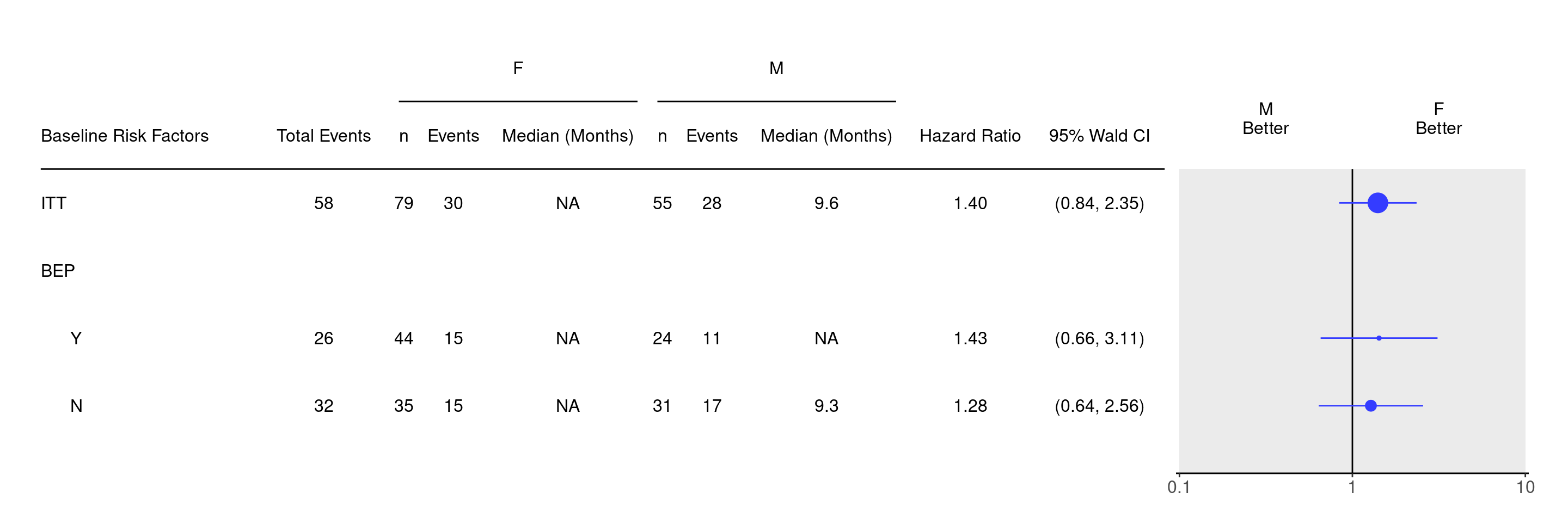
We prepare the data similarly as in SFG3. Additionally we are filtering random.cdisc.data::cadtte to keep only two categories for the SEX variable (otherwise we would not be able to do the forest plot), and we are keeping all ITT patients. We then tabulate statistics to be able to use them as an input for the forest plot.
Code
adtte_mf <- random.cdisc.data::cadtte %>%
df_explicit_na() %>%
filter(
PARAMCD == "OS",
ARM == "A: Drug X",
SEX %in% c("M", "F")
) %>%
droplevels() %>%
mutate(
AVAL = day2month(AVAL),
AVALU = "Months",
is_event = CNSR == 0
) %>%
var_relabel(
BEP01FL = "BEP",
BMRKR1 = "Biomarker (Countinuous)"
)
tbl <- extract_survival_subgroups(
variables = list(
tte = "AVAL",
is_event = "is_event",
arm = "SEX",
subgroups = "BEP01FL"
),
label_all = "ITT",
data = adtte_mf
)
result <- basic_table() %>%
tabulate_survival_subgroups(
df = tbl,
vars = c("n_tot_events", "n", "n_events", "median", "hr", "ci"),
time_unit = adtte_mf$AVALU[1]
)We can now produce the forest plot using the g_forest() function.
R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_1.1.4 tern_0.9.7 rtables_0.6.11 magrittr_2.0.3
[5] formatters_0.5.10
loaded via a namespace (and not attached):
[1] Matrix_1.7-2 gtable_0.3.6 jsonlite_1.9.0
[4] compiler_4.4.2 tidyselect_1.2.1 stringr_1.5.1
[7] tidyr_1.3.1 splines_4.4.2 scales_1.3.0
[10] yaml_2.3.10 fastmap_1.2.0 lattice_0.22-6
[13] ggplot2_3.5.1 R6_2.6.1 labeling_0.4.3
[16] generics_0.1.3 knitr_1.49 forcats_1.0.0
[19] rbibutils_2.3 htmlwidgets_1.6.4 backports_1.5.0
[22] checkmate_2.3.2 tibble_3.2.1 munsell_0.5.1
[25] pillar_1.10.1 rlang_1.1.5 broom_1.0.7
[28] stringi_1.8.4 xfun_0.51 cli_3.6.4
[31] withr_3.0.2 Rdpack_2.6.2 digest_0.6.37
[34] grid_4.4.2 cowplot_1.1.3 lifecycle_1.0.4
[37] vctrs_0.6.5 evaluate_1.0.3 glue_1.8.0
[40] farver_2.1.2 nestcolor_0.1.3 codetools_0.2-20
[43] survival_3.8-3 random.cdisc.data_0.3.16 colorspace_2.1-1
[46] rmarkdown_2.29 purrr_1.0.4 tools_4.4.2
[49] pkgconfig_2.0.3 htmltools_0.5.8.1 Reuse
Copyright 2023, Hoffmann-La Roche Ltd.