---
title: SFG5B
subtitle: Survival Forest Graph Comparing Categorical Biomarker Groups on Subgroups, including ITT
categories: [SFG]
---
------------------------------------------------------------------------
::: panel-tabset
{{< include setup.qmd >}}
## Plot
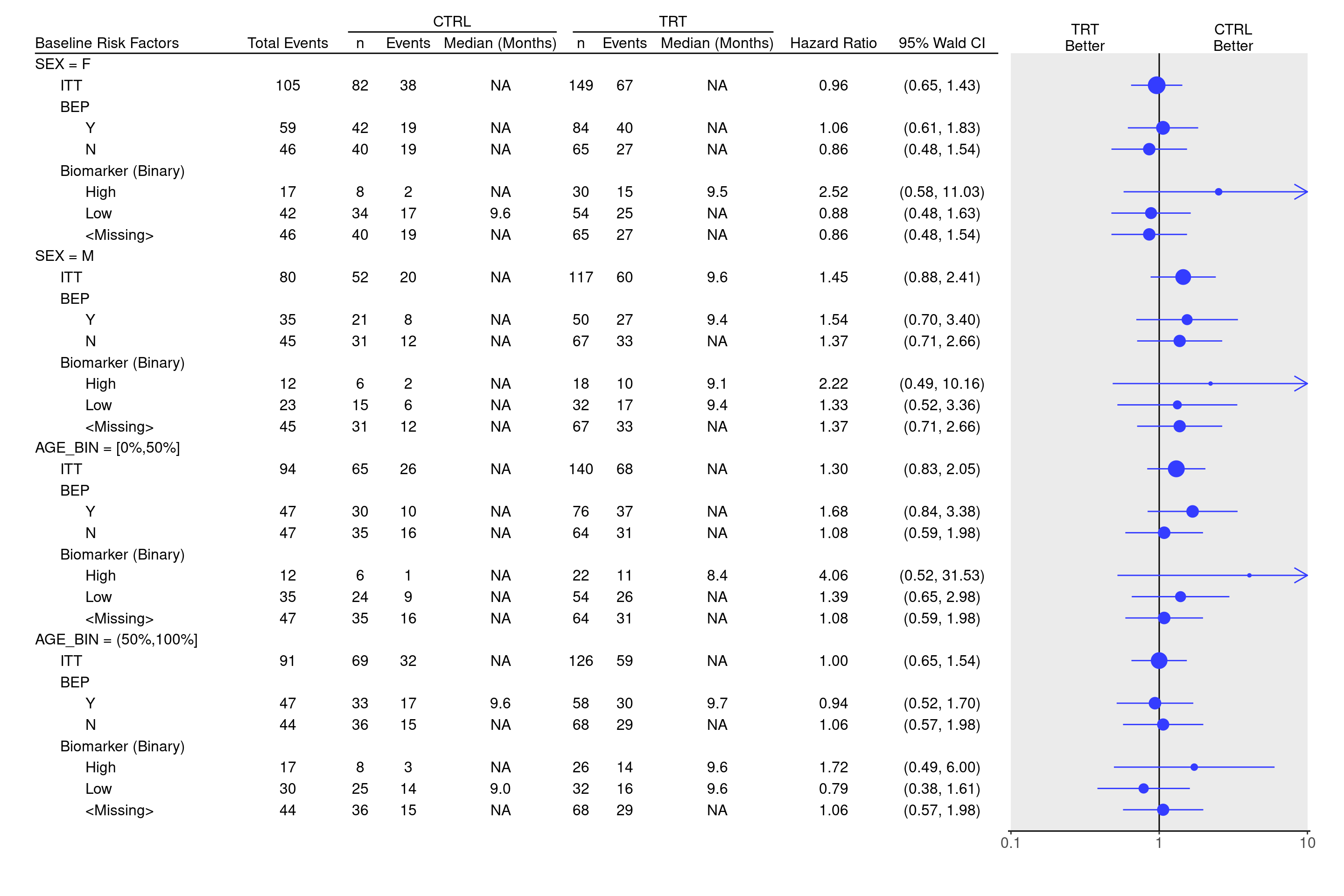
We use the same `tables_all()` function above and combine all subtables using `rbind()` to tabulate statistics to be able to use as an input for forest plot.
```{r}
tables_all <- function(filter_var, filter_condition, sub_var) {
dataset <- adtte %>%
filter(!!as.name(filter_var) == filter_condition)
if (nrow(dataset) == 0) {
stop(paste("Subset", filter_var, "==", filter_condition, "is empty"))
}
tbl <- extract_survival_subgroups(
variables = list(
tte = "AVAL",
is_event = "is_event",
arm = "ARM_BIN",
subgroups = sub_var
),
label_all = "ITT",
data = dataset
)
basic_table() %>%
tabulate_survival_subgroups(
df = tbl,
vars = c("n_tot_events", "n", "n_events", "median", "hr", "ci"),
time_unit = dataset$AVALU[1]
)
}
add_subtitle <- function(sub_tab, sub_title) {
label_at_path(sub_tab, path = row_paths(sub_tab)[[1]][1]) <- sub_title
sub_tab
}
tables_list <- list(
tables_all(filter_var = "SEX", filter_condition = "F", sub_var = c("BEP01FL", "BMRKR2_BIN")),
tables_all(filter_var = "SEX", filter_condition = "M", sub_var = c("BEP01FL", "BMRKR2_BIN")),
tables_all(filter_var = "AGE_BIN", filter_condition = "[0%,50%]", sub_var = c("BEP01FL", "BMRKR2_BIN")),
tables_all(filter_var = "AGE_BIN", filter_condition = "(50%,100%]", sub_var = c("BEP01FL", "BMRKR2_BIN"))
)
```
We can now produce the forest plot using the `g_forest()` function.
```{r, fig.width = 15, fig.height = 10}
one_table <- tables_list[[1]]
result <- rbind(
add_subtitle(tables_list[[1]], "SEX = F"),
add_subtitle(tables_list[[2]], "SEX = M"),
add_subtitle(tables_list[[3]], "AGE_BIN = [0%,50%]"),
add_subtitle(tables_list[[4]], "AGE_BIN = (50%,100%]")
)
g_forest(
result,
col_x = attr(one_table, "col_x"),
col_ci = attr(one_table, "col_ci"),
forest_header = attr(one_table, "forest_header"),
col_symbol_size = attr(one_table, "col_symbol_size")
)
```
{{< include ../../misc/session_info.qmd >}}
:::