DG2
Boxplots of a Numeric Variable by Categorical Variables
The graph below plots the distribution of a biomarker variable (on a continuous scale) as a boxplot by one or more categorical clinical variables with overlaid points.
We will use the cadsl data set from the random.cdisc.data package to illustrate the graph and will select the biomarker evaluable population with BEP01FL. The column BMRKR1 contains the biomarker values on a continuous scale. We will use STRATA2 and ARM as categorical clinical variables.
Here below the code for a simple boxplot with the outliers displayed. Note that you may run into warning messages after producing the graph if the variable you want to analyze contains NAs. To avoid these warning messages, you can use the drop_na() function from tidyr in the data manipulation step above to remove the NAs rows from the dataset (e.g drop_na(BMRKR1)).
Code
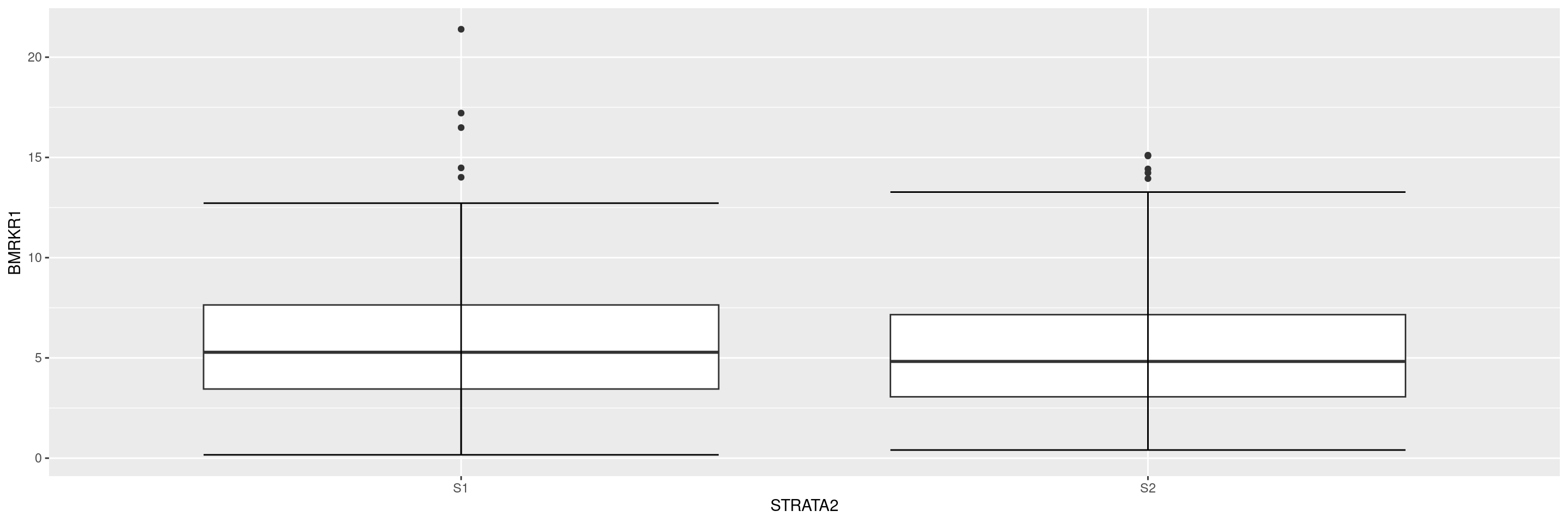
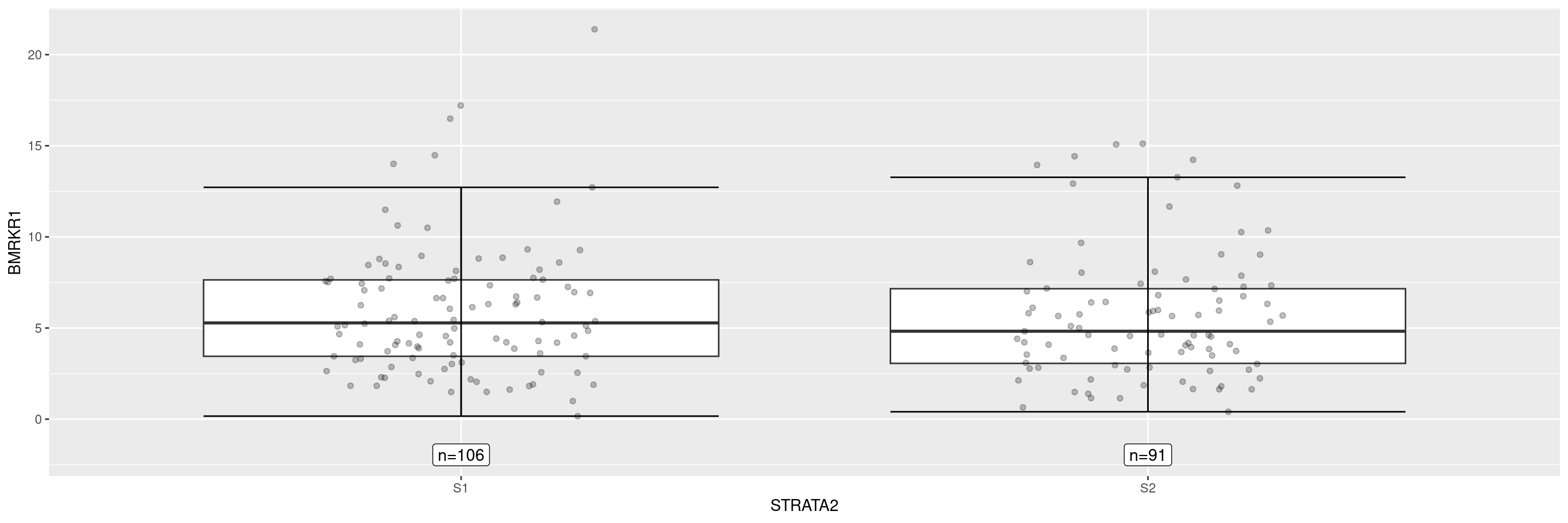
Now we overlay the original data points, and remove the display of the outliers to avoid duplicate points.
Code
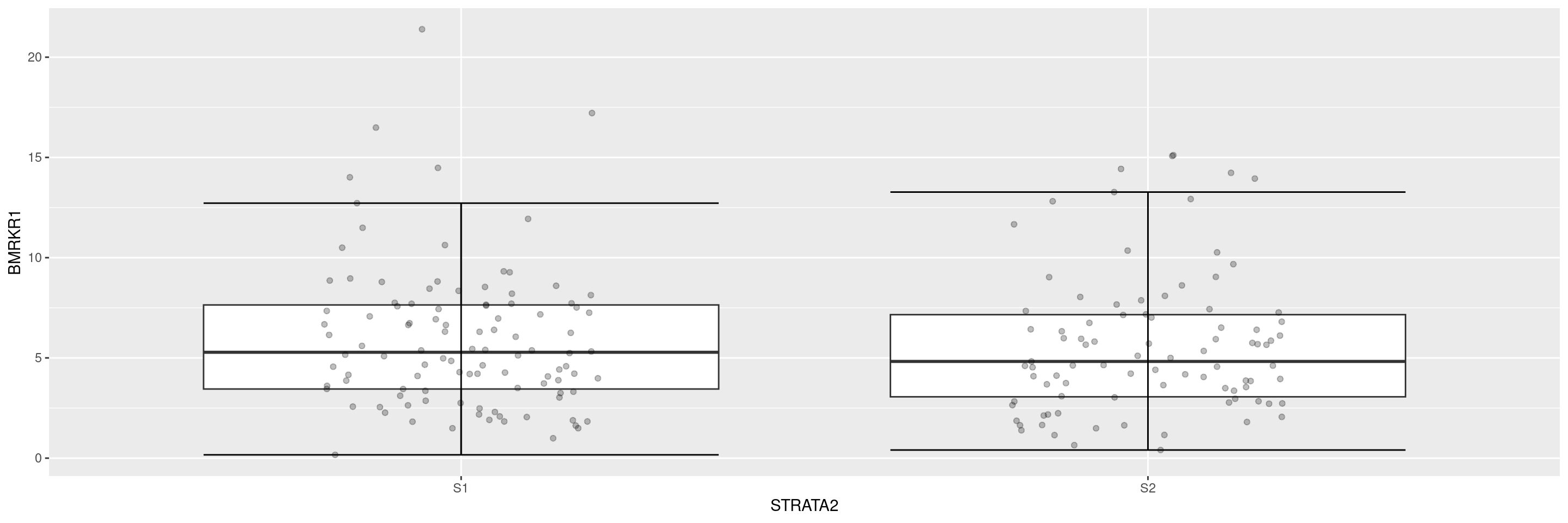
We can customize the labels of the axes.
Code
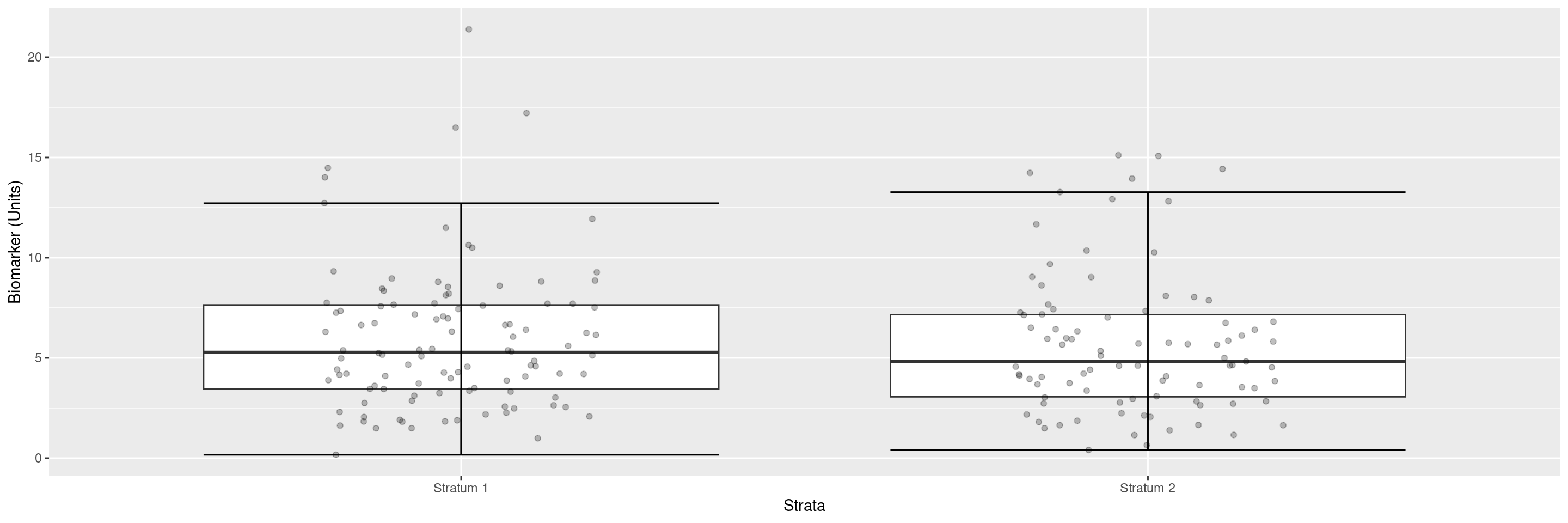
We can add the group sizes as annotations.
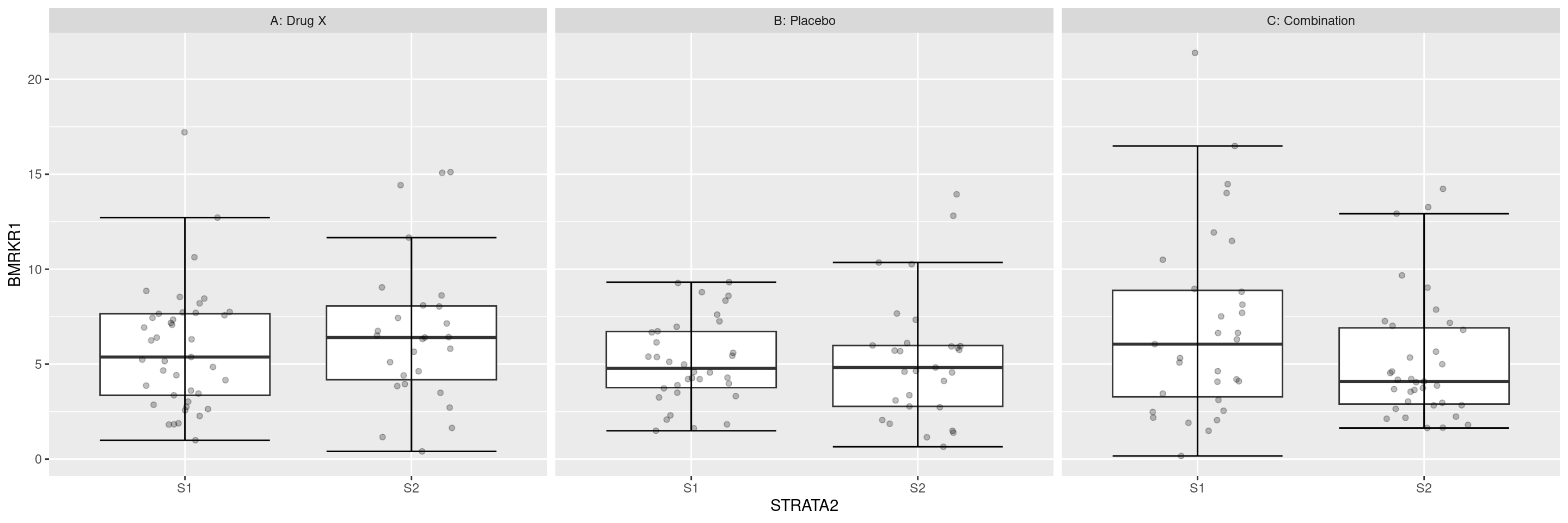
We can also display the biomarker by a further categorical variable with the facet_grid() layer.
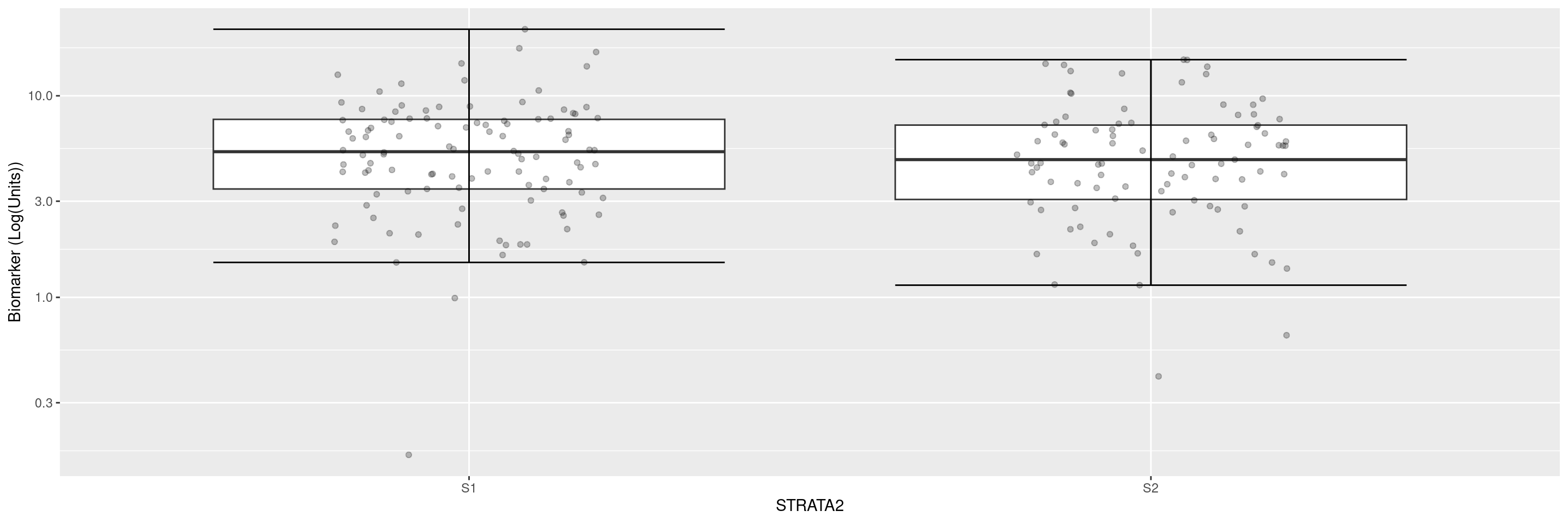
This example shows how to display the biomarker axis on a log scale.
R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_1.1.4 ggplot2.utils_0.3.2 ggplot2_3.5.1
[4] tern_0.9.7 rtables_0.6.11 magrittr_2.0.3
[7] formatters_0.5.10
loaded via a namespace (and not attached):
[1] generics_0.1.3 tidyr_1.3.1 EnvStats_3.0.0
[4] stringi_1.8.4 lattice_0.22-6 digest_0.6.37
[7] evaluate_1.0.3 grid_4.4.2 fastmap_1.2.0
[10] jsonlite_1.9.0 Matrix_1.7-2 backports_1.5.0
[13] survival_3.8-3 purrr_1.0.4 scales_1.3.0
[16] codetools_0.2-20 Rdpack_2.6.2 cli_3.6.4
[19] ggpp_0.5.8-1 nestcolor_0.1.3 rlang_1.1.5
[22] rbibutils_2.3 munsell_0.5.1 splines_4.4.2
[25] withr_3.0.2 yaml_2.3.10 tools_4.4.2
[28] polynom_1.4-1 checkmate_2.3.2 colorspace_2.1-1
[31] forcats_1.0.0 ggstats_0.8.0 broom_1.0.7
[34] vctrs_0.6.5 R6_2.6.1 lifecycle_1.0.4
[37] stringr_1.5.1 htmlwidgets_1.6.4 MASS_7.3-64
[40] pkgconfig_2.0.3 pillar_1.10.1 gtable_0.3.6
[43] glue_1.8.0 xfun_0.51 tibble_3.2.1
[46] tidyselect_1.2.1 knitr_1.49 farver_2.1.2
[49] htmltools_0.5.8.1 labeling_0.4.3 rmarkdown_2.29
[52] random.cdisc.data_0.3.16 compiler_4.4.2