KG1A
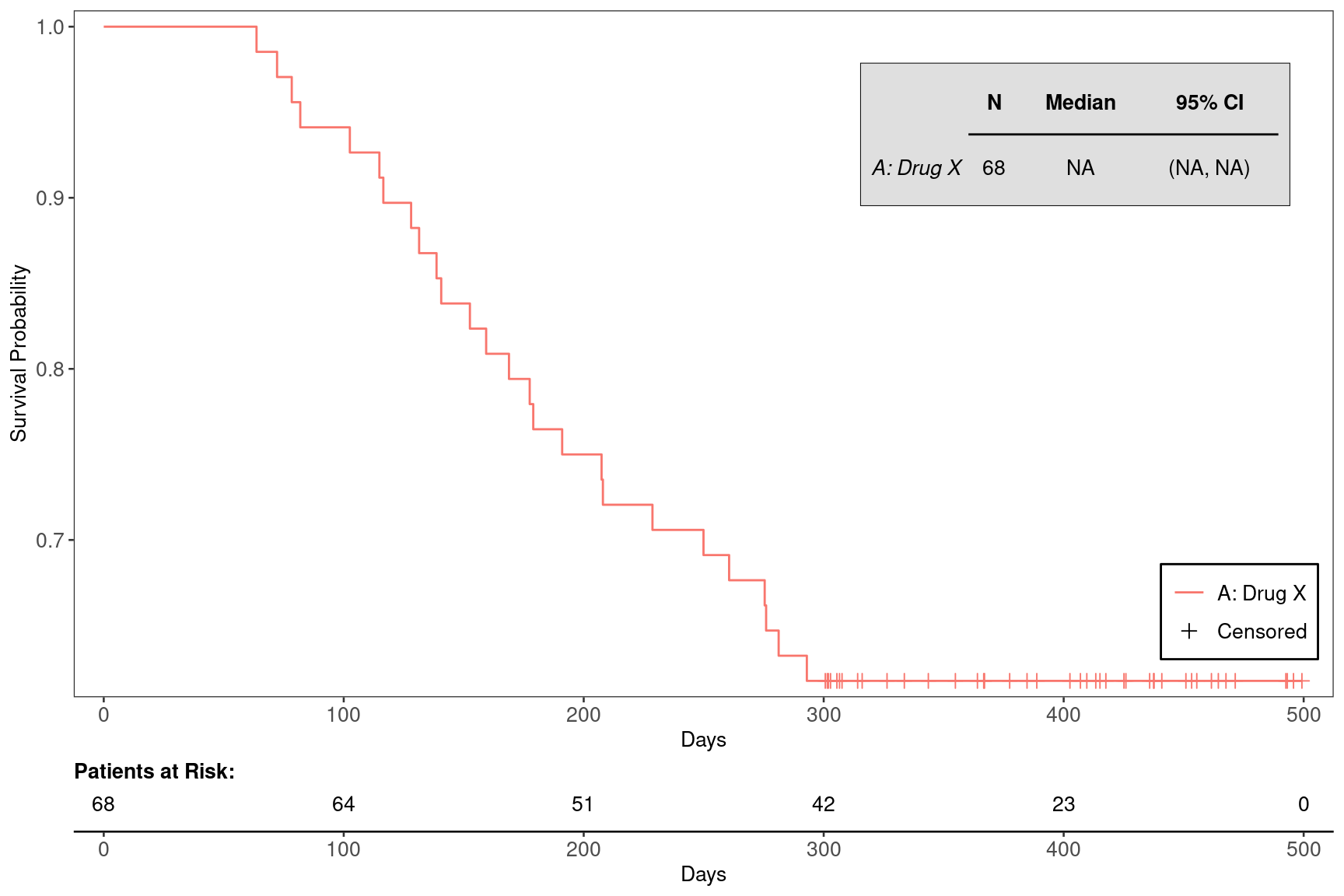
Kaplan-Meier Graph for Biomarker Evaluable Population in One Treatment Arm
KG
We will use the cadtte data set from the random.cdisc.data package to create the Kaplan-Meier (KM) plots. We start by filtering the time-to-event dataset for the overall survival observations and by one treatment arm (A), creating a new variable for event information, and curating a list of variables required to produce the plot.
We can filter the dataset further for the biomarker evaluable population using the corresponding flag variable, here BEP01FL:
Afterwards we can plot the basic KM graph, just using the further filtered dataset adtte_bep. Here we annotate the plot with median survival time, but could suppress it with annot_surv_med = FALSE.
R version 4.4.2 (2024-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] ggplot2_3.5.1 dplyr_1.1.4 tern_0.9.7 rtables_0.6.11
[5] magrittr_2.0.3 formatters_0.5.10
loaded via a namespace (and not attached):
[1] Matrix_1.7-2 gtable_0.3.6 jsonlite_1.9.0
[4] compiler_4.4.2 tidyselect_1.2.1 stringr_1.5.1
[7] tidyr_1.3.1 splines_4.4.2 scales_1.3.0
[10] yaml_2.3.10 fastmap_1.2.0 lattice_0.22-6
[13] R6_2.6.1 labeling_0.4.3 generics_0.1.3
[16] knitr_1.49 forcats_1.0.0 rbibutils_2.3
[19] htmlwidgets_1.6.4 backports_1.5.0 checkmate_2.3.2
[22] tibble_3.2.1 munsell_0.5.1 pillar_1.10.1
[25] rlang_1.1.5 broom_1.0.7 stringi_1.8.4
[28] xfun_0.51 cli_3.6.4 withr_3.0.2
[31] Rdpack_2.6.2 digest_0.6.37 cowplot_1.1.3
[34] lifecycle_1.0.4 vctrs_0.6.5 evaluate_1.0.3
[37] glue_1.8.0 farver_2.1.2 nestcolor_0.1.3
[40] codetools_0.2-20 survival_3.8-3 random.cdisc.data_0.3.16
[43] colorspace_2.1-1 rmarkdown_2.29 purrr_1.0.4
[46] tools_4.4.2 pkgconfig_2.0.3 htmltools_0.5.8.1 Reuse
Copyright 2023, Hoffmann-La Roche Ltd.