---
title: AET09_SMQ
subtitle: Adverse Events Related to Study Drug by Standardized MedDRA Query
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(dplyr)
library(tern)
library(stringr)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing
# levels. For details, refer to Teal and Study Data article.
adsl <- df_explicit_na(adsl)
adae <- df_explicit_na(adae)
# AEs are filtered for relatedness to study drug.
adae_rel <- adae %>% filter(AEREL == "Y")
# Simulate a random AAG dataset.
aag <- data.frame(
NAMVAR = c("SMQ01NAM", "SMQ01NAM", "SMQ02NAM", "CQ01NAM", "CQ01NAM"),
SRCVAR = rep("AEDECOD", 5),
GRPTYPE = c("SMQ", "SMQ", "SMQ", "CUSTOM", "CUSTOM"),
REFID = c(1, 1, 2, 3, 3),
REFNAME = c(rep("C.1.1.1.3/B.2.2.3.1 AESI", 2), "non observed SMQ02NAM level", rep("D.2.1.5.3/A.1.1.1.1 AESI", 2)),
SCOPE = c("BROAD", "BROAD", "BROAD", "", ""),
REFTERM = c("C.1.1.1.3", "B.2.2.3.1", "Z.9.9.9.9", "D.2.1.5.3", "A.1.1.1.1"),
stringsAsFactors = FALSE
)
# Create summary AAG dataset (used for labelling).
# Note it's important to incorporate SCOPE into the basket names for SMQs so as to
# match the flags present in ADAE.
aag_summary <- aag %>%
select(NAMVAR, REFNAME, SCOPE) %>%
unique() %>%
mutate(
REFNAME_SCOPE = ifelse(SCOPE == "", REFNAME, paste0(REFNAME, "(", SCOPE, ")"))
) %>%
rename(basket = NAMVAR, basket_name = REFNAME_SCOPE) %>%
select(basket, basket_name)
# Make a summary of the full ADAE based on AAG by using h_stack_by_baskets helper function
adae_smq_all <- h_stack_by_baskets(
df = adae_rel,
aag_summary = aag_summary,
keys = c("STUDYID", "USUBJID", "ACTARMCD", "AEDECOD")
)
# Post-process adae_smq_all to keep only certain baskets of interest for variant 1
# Not need to drop baskets for variant 2 as all baskets will be considered.
baskets_to_drop_variant <- aag_summary$basket_name[!aag_summary$basket %in% c("SMQ01NAM")]
# Adjust the analysis dataset based on basket subset for variant 1.
adae_smq_1 <- adae_smq_all %>%
filter(!adae_smq_all$SMQ %in% baskets_to_drop_variant) %>%
mutate(
SMQ = tern::fct_discard(SMQ, discard = baskets_to_drop_variant)
)
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
::::: panel-tabset
## Standard Table
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result")}
criteria_fun <- function(tr) !is(tr, "ContentRow") && all_zero_or_na(tr)
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARMCD") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique"),
.labels = c(
unique = "Total number of patients with at least one adverse event"
)
) %>%
split_rows_by(
"SMQ",
child_labels = "visible",
nested = FALSE,
split_fun = trim_levels_in_group("AEDECOD", drop_outlevs = FALSE),
label_pos = "topleft",
split_label = "Standardized MedDRA Query"
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Total number of events"
)
) %>%
count_occurrences(
vars = "AEDECOD",
.indent_mods = -1L,
drop = FALSE
) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
result <- build_table(
lyt,
df = adae_smq_1,
alt_counts_df = adsl
) %>%
sort_at_path(path = c("SMQ"), scorefun = cont_n_allcols) %>%
sort_at_path(path = c("SMQ", "*", "AEDECOD"), scorefun = score_occurrences, na.pos = "last") %>%
trim_rows(criteria = criteria_fun)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Customized Queries
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant2, test = list(result_v2 = "result")}
criteria_fun <- function(tr) {
!is(tr, "ContentRow") && all_zero_or_na(tr) && !grepl("Total number of", obj_label(tr))
}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARMCD") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique"),
.labels = c(
unique = "Total number of patients with at least one adverse event"
)
) %>%
split_rows_by(
"SMQ",
child_labels = "visible",
nested = FALSE,
split_fun = trim_levels_in_group("AEDECOD", drop_outlevs = FALSE),
label_pos = "topleft",
split_label = "Standardized MedDRA Query"
) %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Total number of events"
),
show_labels = "hidden"
) %>%
count_occurrences(
vars = "AEDECOD",
drop = FALSE
) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
score_ae_pts <- function(tt) {
count_vals <- cell_values(tree_children(tt)[["USUBJID"]])[[1]]
sum(matrix(unlist(count_vals), nrow = length(count_vals), byrow = TRUE)[, 1])
}
result <- build_table(
lyt,
df = adae_smq_all,
alt_counts_df = adsl
) %>%
sort_at_path(path = c("SMQ"), scorefun = score_ae_pts) %>%
sort_at_path(path = c("SMQ", "*", "AEDECOD"), scorefun = score_occurrences, na.pos = "last") %>%
trim_rows(criteria = criteria_fun)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
:::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
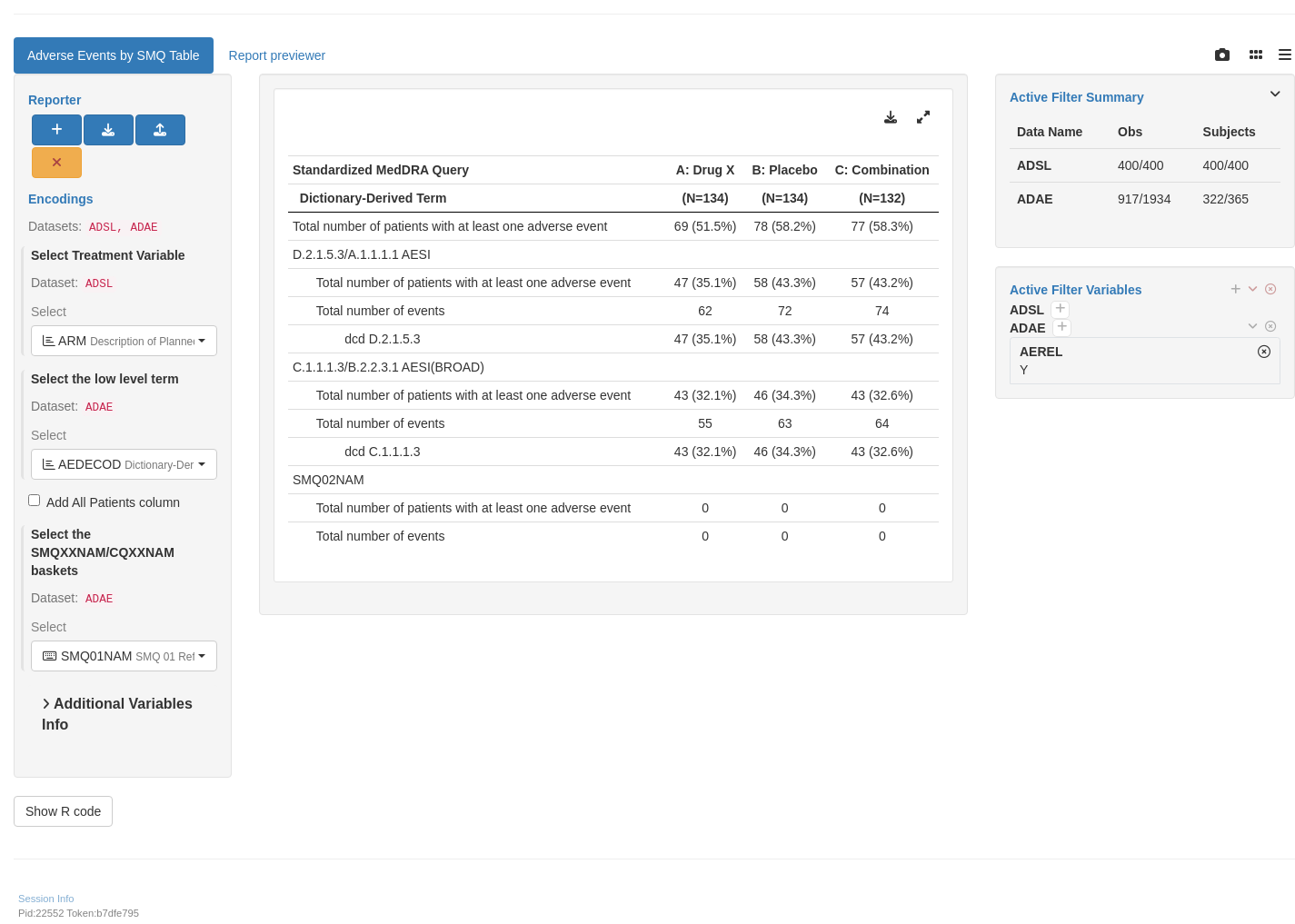
Note that filtering AEs for relatedness to study drug is not a necessary data pre-processing step for the module. It can be achieved using the `teal` module filter panel. The example here pre-sets the filters using `AEREL = "Y"` in `adae`.
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
ADSL <- random.cdisc.data::cadsl
ADAE <- random.cdisc.data::cadae
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADAE")]
## Reusable Configuration For Modules
ADSL <- data[["ADSL"]]
ADAE <- data[["ADAE"]]
names_baskets <- grep("^(SMQ|CQ).*NAM$", names(ADAE), value = TRUE)
names_scopes <- grep("^SMQ.*SC$", names(ADAE), value = TRUE)
cs_baskets <- choices_selected(
choices = variable_choices(ADAE, subset = names_baskets),
selected = names_baskets
)
cs_scopes <- choices_selected(
choices = variable_choices(ADAE, subset = names_scopes),
selected = names_scopes,
fixed = TRUE
)
## Setup App
app <- init(
data = data,
modules = modules(
tm_t_smq(
label = "Adverse Events by SMQ Table",
dataname = "ADAE",
arm_var = choices_selected(
choices = variable_choices(ADSL, subset = c("ARM", "SEX")),
selected = "ARM"
),
add_total = FALSE,
baskets = cs_baskets,
scopes = cs_scopes,
llt = choices_selected(
choices = variable_choices(ADAE, subset = c("AEDECOD")),
selected = "AEDECOD"
)
)
),
filter = teal_slices(teal_slice("ADAE", "AEREL", selected = "Y"))
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}