---
title: EGT05_QTCAT
subtitle: ECG Actual Values and Changes from Baseline by Visit
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(tern)
library(dplyr)
adsl <- random.cdisc.data::cadsl
adeg <- random.cdisc.data::cadeg
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
adsl <- df_explicit_na(adsl)
adeg <- df_explicit_na(adeg)
adeg_labels <- var_labels(adeg)
adeg_f <- adeg %>%
filter(
PARAMCD == "QT",
ANL01FL == "Y"
) %>%
mutate(
AVALCAT1 = case_when(
AVAL <= 450 ~ "<=450 msec",
AVAL <= 480 ~ ">450 to <=480 msec",
AVAL <= 500 ~ ">480 to <= 500 msec",
AVAL > 500 ~ ">500 msec",
is.na(AVAL) ~ "<Missing>"
),
CHGCAT1 = case_when(
CHG <= 30 ~ "<=30 msec",
CHG <= 60 ~ ">30 to <=60 msec",
CHG > 60 ~ ">60 msec",
is.na(CHG) ~ "<Missing>"
)
) %>%
mutate(
AVALCAT1 = factor(
AVALCAT1,
levels = c(
"<=450 msec",
">450 to <=480 msec",
">480 to <= 500 msec",
">500 msec",
"<Missing>"
)
),
CHGCAT1 = factor(
CHGCAT1,
levels = c(
"<=30 msec",
">30 to <=60 msec",
">60 msec",
"<Missing>"
)
)
) %>%
var_relabel(
AVALCAT1 = "Value at Visit",
CHGCAT1 = "Change from Baseline"
)
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
:::: panel-tabset
## Standard Table
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
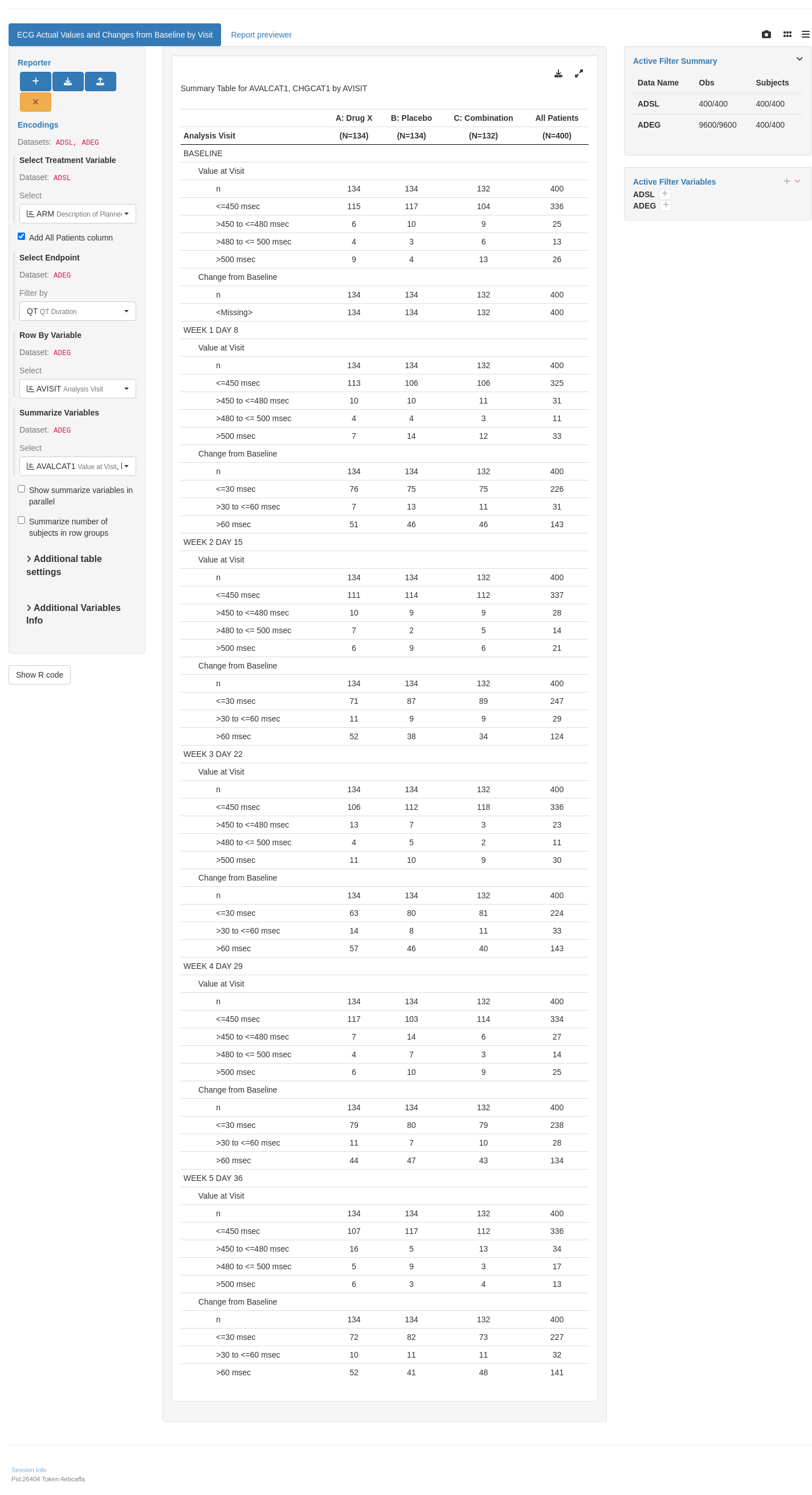
```{r variant1, test = list(result_v1 = "result")}
split_fun <- drop_split_levels
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ARM") %>%
split_rows_by(
"PARAM",
split_label = obj_label(adeg_f$PARAM),
split_fun = split_fun,
label_pos = "topleft"
) %>%
split_rows_by(
"AVISIT",
split_label = obj_label(adeg_f$AVISIT),
split_fun = split_fun,
label_pos = "topleft"
) %>%
analyze_vars(
vars = c("AVALCAT1", "CHGCAT1"),
var_labels = c("Value at Visit", "Change from Baseline")
) %>%
append_topleft(" Category")
result <- build_table(lyt = lyt, df = adeg_f, alt_counts_df = adsl) %>%
prune_table()
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
library(dplyr)
ADSL <- random.cdisc.data::cadsl
ADEG <- random.cdisc.data::cadeg
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
ADSL <- df_explicit_na(ADSL)
ADEG <- df_explicit_na(ADEG)
adeg_labels <- col_labels(ADEG)
ADEG <- ADEG %>%
filter(
ANL01FL == "Y" # no need to filter for PARAMCD here
) %>%
mutate(
AVALCAT1 = case_when(
AVAL <= 450 ~ "<=450 msec",
AVAL <= 480 ~ ">450 to <=480 msec",
AVAL <= 500 ~ ">480 to <= 500 msec",
AVAL > 500 ~ ">500 msec",
is.na(AVAL) ~ "<Missing>"
),
CHGCAT1 = case_when(
CHG <= 30 ~ "<=30 msec",
CHG <= 60 ~ ">30 to <=60 msec",
CHG > 60 ~ ">60 msec",
is.na(CHG) ~ "<Missing>"
)
) %>%
mutate(
AVALCAT1 = factor(
AVALCAT1,
levels = c(
"<=450 msec",
">450 to <=480 msec",
">480 to <= 500 msec",
">500 msec",
"<Missing>"
)
),
CHGCAT1 = factor(
CHGCAT1,
levels = c(
"<=30 msec",
">30 to <=60 msec",
">60 msec",
"<Missing>"
)
)
)
col_labels(ADEG) <- c(
adeg_labels,
"AVALCAT1" = "Value at Visit",
"CHGCAT1" = "Change from Baseline"
)
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADEG")]
## Reusable Configuration For Modules
ADSL <- data[["ADSL"]]
ADEG <- data[["ADEG"]]
## Setup App
app <- init(
data = data,
modules = modules(
tm_t_summary_by(
label = "ECG Actual Values and Changes from Baseline by Visit",
dataname = "ADEG",
arm_var = choices_selected(
choices = variable_choices(ADSL, c("ARM", "ARMCD")),
selected = "ARM"
),
by_vars = choices_selected(
choices = variable_choices(ADEG, c("PARAM", "AVISIT")),
selected = c("AVISIT")
),
summarize_vars = choices_selected(
choices = variable_choices(ADEG, c("AVALCAT1", "CHGCAT1")),
selected = c("AVALCAT1", "CHGCAT1")
),
useNA = "ifany",
paramcd = choices_selected(
choices = value_choices(ADEG, "PARAMCD", "PARAM"),
selected = "QT"
)
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}