---
title: AET01
subtitle: Overview of Deaths and Adverse Events
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(tern)
library(dplyr)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
adsl <- df_explicit_na(adsl)
adae <- df_explicit_na(
adae,
omit_columns = c("SMQ01NAM", "SMQ01SC", "SMQ02NAM", "SMQ02SC", "CQ01NAM", "STUDYID", "USUBJID")
)
set.seed(99)
adae <- adae %>%
mutate(
AEDECOD = with_label(as.character(AEDECOD), "Dictionary-Derived Term"),
AESDTH = with_label(
sample(c("N", "Y"), size = nrow(adae), replace = TRUE, prob = c(0.99, 0.01)),
"Results in Death"
),
AEACN = with_label(
sample(
c("DOSE NOT CHANGED", "DOSE INCREASED", "DRUG INTERRUPTED", "DRUG WITHDRAWN"),
size = nrow(adae),
replace = TRUE, prob = c(0.68, 0.02, 0.25, 0.05)
),
"Action Taken with Study Treatment"
),
FATAL = with_label(AESDTH == "Y", "AE with fatal outcome"),
SEV = with_label(AESEV == "SEVERE", "Severe AE (at greatest intensity)"),
SER = with_label(AESER == "Y", "Serious AE"),
SERWD = with_label(AESER == "Y" & AEACN == "DRUG WITHDRAWN", "Serious AE leading to withdrawal from treatment"),
SERDSM = with_label(
AESER == "Y" & AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"Serious AE leading to dose modification/interruption"
),
RELSER = with_label(AESER == "Y" & AEREL == "Y", "Related Serious AE"),
WD = with_label(AEACN == "DRUG WITHDRAWN", "AE leading to withdrawal from treatment"),
DSM = with_label(
AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"), "AE leading to dose modification/interruption"
),
REL = with_label(AEREL == "Y", "Related AE"),
RELWD = with_label(AEREL == "Y" & AEACN == "DRUG WITHDRAWN", "Related AE leading to withdrawal from treatment"),
RELDSM = with_label(
AEREL == "Y" & AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"Related AE leading to dose modification/interruption"
),
CTC35 = with_label(AETOXGR %in% c("3", "4", "5"), "Grade 3-5 AE"),
CTC45 = with_label(AETOXGR %in% c("4", "5"), "Grade 4/5 AE"),
SMQ01 = with_label(SMQ01NAM != "", aesi_label(adae$SMQ01NAM, adae$SMQ01SC)),
SMQ02 = with_label(SMQ02NAM != "", aesi_label(adae$SMQ02NAM, adae$SMQ02SC)),
CQ01 = with_label(CQ01NAM != "", aesi_label(adae$CQ01NAM)),
USUBJID_AESEQ = paste(USUBJID, AESEQ, sep = "@@") # Create unique ID per AE in dataset.
) %>%
filter(ANL01FL == "Y")
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
::::::: panel-tabset
## Standard Table
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result")}
aesi_vars <- c("FATAL", "SER", "SERWD", "SERDSM", "RELSER", "WD", "DSM", "REL", "RELWD", "RELDSM", "SEV")
# Layout for variables from adsl dataset.
lyt_adsl <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
count_patients_with_event(
"USUBJID",
filters = c("DTHFL" = "Y"),
denom = "N_col",
.labels = c(count_fraction = "Total number of deaths")
) %>%
count_patients_with_event(
"USUBJID",
filters = c("DCSREAS" = "ADVERSE EVENT"),
denom = "N_col",
.labels = c(count_fraction = "Total number of patients withdrawn from study due to an AE"),
table_names = "tot_wd"
)
result_adsl <- build_table(lyt_adsl, df = adsl, alt_counts_df = adsl)
# Layout for variables from adae dataset.
lyt_adae <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one AE",
nonunique = "Total number of AEs"
),
.formats = list(unique = format_count_fraction_fixed_dp, nonunique = "xx"),
show_labels = "hidden"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_vars,
denom = "N_col",
var_labels = "Total number of patients with at least one",
show_labels = "visible"
)
result_adae <- build_table(lyt_adae, df = adae, alt_counts_df = adsl)
# Combine tables.
col_info(result_adsl) <- col_info(result_adae)
result <- rbind(
result_adae[1:2, ],
result_adsl,
result_adae[3:nrow(result_adae), ]
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Medical <br/> Concepts Section
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant2, test = list(result_v2 = "result")}
aesi_vars <- c("FATAL", "SER", "SERWD", "SERDSM", "RELSER", "WD", "DSM", "REL", "RELWD", "RELDSM", "CTC35")
basket_vars <- c("SMQ01", "SMQ02", "CQ01")
# Layout for variables from adsl dataset.
lyt_adsl <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
count_patients_with_event(
"USUBJID",
filters = c("DTHFL" = "Y"),
denom = "N_col",
.labels = c(count_fraction = "Total number of deaths")
) %>%
count_patients_with_event(
"USUBJID",
filters = c("DCSREAS" = "ADVERSE EVENT"),
denom = "N_col",
.labels = c(count_fraction = "Total number of patients withdrawn from study due to an AE"),
table_names = "tot_wd"
)
result_adsl <- build_table(lyt_adsl, df = adsl, alt_counts_df = adsl)
# Layout for variables from adae dataset.
lyt_adae <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one AE",
nonunique = "Total number of AEs"
),
.formats = list(unique = format_count_fraction_fixed_dp, nonunique = "xx"),
show_labels = "hidden"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_vars,
denom = "N_col",
var_labels = "Total number of patients with at least one",
show_labels = "visible"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = basket_vars,
table_names = "table_aesi",
denom = "N_col",
var_labels = "Total number of patients with at least one",
show_labels = "visible"
)
result_adae <- build_table(lyt_adae, df = adae, alt_counts_df = adsl)
# Combine tables.
col_info(result_adsl) <- col_info(result_adae)
result <- rbind(
result_adae[1:2, ],
result_adsl,
result_adae[3:nrow(result_adae), ]
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with <br/> Modified Rows
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant3, test = list(result_v3 = "result")}
aesi_vars <- c("FATAL", "SER", "WD", "REL", "CTC35", "CTC45")
# Layout for variables from adsl dataset.
lyt_adsl <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
count_patients_with_event(
"USUBJID",
filters = c("DTHFL" = "Y"),
denom = "N_col",
.labels = c(count_fraction = "Total number of deaths")
) %>%
count_patients_with_event(
"USUBJID",
filters = c("DCSREAS" = "ADVERSE EVENT"),
denom = "N_col",
.labels = c(count_fraction = "Total number of patients withdrawn from study due to an AE"),
table_names = "tot_wd"
) %>%
count_patients_with_event(
"USUBJID",
filters = c("DCSREAS" = "WITHDRAWAL BY SUBJECT"),
denom = "N_col",
.labels = c(count_fraction = "Total number of patients withdrawn informed consent"),
table_names = "tot_dscsreas_wd"
)
result_adsl <- build_table(lyt_adsl, df = adsl, alt_counts_df = adsl)
# Layout for variables from adae dataset.
lyt_adae <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one AE",
nonunique = "Total number of AEs"
),
.formats = list(unique = format_count_fraction_fixed_dp, nonunique = "xx"),
show_labels = "hidden"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = aesi_vars,
denom = "N_col",
var_labels = "Total number of patients with at least one",
show_labels = "visible"
)
result_adae <- build_table(lyt_adae, df = adae, alt_counts_df = adsl)
# Combine tables.
col_info(result_adsl) <- col_info(result_adae)
result <- rbind(
result_adae[1:2, ],
result_adsl,
result_adae[3:nrow(result_adae), ]
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant3")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Rows Counting <br/> Events & Additional Sections
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant4, test = list(result_v4 = "result")}
count_subj_vars <- c("FATAL", "SER", "WD", "DSM", "REL", "CTC35")
count_term_vars <- c("SER", "DSM", "REL", "CTC35", "CTC45")
count_ae_vars <- c("SER", "DSM", "REL", "CTC35", "CTC45")
# Layout for variables from adsl dataset.
lyt_adsl <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
count_patients_with_event(
"USUBJID",
filters = c("DTHFL" = "Y"),
denom = "N_col",
.labels = c(count_fraction = "Total number of deaths")
) %>%
count_patients_with_event(
"USUBJID",
filters = c("DCSREAS" = "ADVERSE EVENT"),
denom = "N_col",
.labels = c(count_fraction = "Total number of patients withdrawn from study due to an AE"),
table_names = "tot_wd"
)
result_adsl <- build_table(lyt_adsl, df = adsl, alt_counts_df = adsl)
# Layout for variables from adae dataset.
lyt_adae <- basic_table(show_colcounts = TRUE) %>%
split_cols_by("ACTARM") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one AE",
nonunique = "Total number of AEs"
),
.formats = list(unique = format_count_fraction_fixed_dp, nonunique = "xx"),
show_labels = "hidden"
) %>%
count_patients_with_flags(
"USUBJID",
flag_variables = count_subj_vars,
denom = "N_col",
var_labels = "Total number of patients with at least one",
show_labels = "visible"
) %>%
count_patients_with_flags(
"AEDECOD",
flag_variables = count_term_vars,
.stats = "count",
.formats = c(count = "xx"),
table_names = "table_term",
var_labels = "Total number of unique preferred terms which are",
show_labels = "visible"
) %>%
count_patients_with_flags(
"USUBJID_AESEQ",
flag_variables = count_ae_vars,
.stats = "count",
.formats = c(count = "xx"),
table_names = "table_ae",
var_labels = "Total number of adverse events which are",
show_labels = "visible"
)
result_adae <- build_table(lyt_adae, df = adae, alt_counts_df = adsl)
# Combine tables.
col_info(result_adsl) <- col_info(result_adae)
result <- rbind(
result_adae[1:2, ],
result_adsl,
result_adae[3:nrow(result_adae), ]
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant4")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
To illustrate, additional variables such as flags (TRUE/FALSE) for select AEs of interest and select AE baskets are added to the `adae` dataset.
```{r setup}
#| code-fold: show
```
:::::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
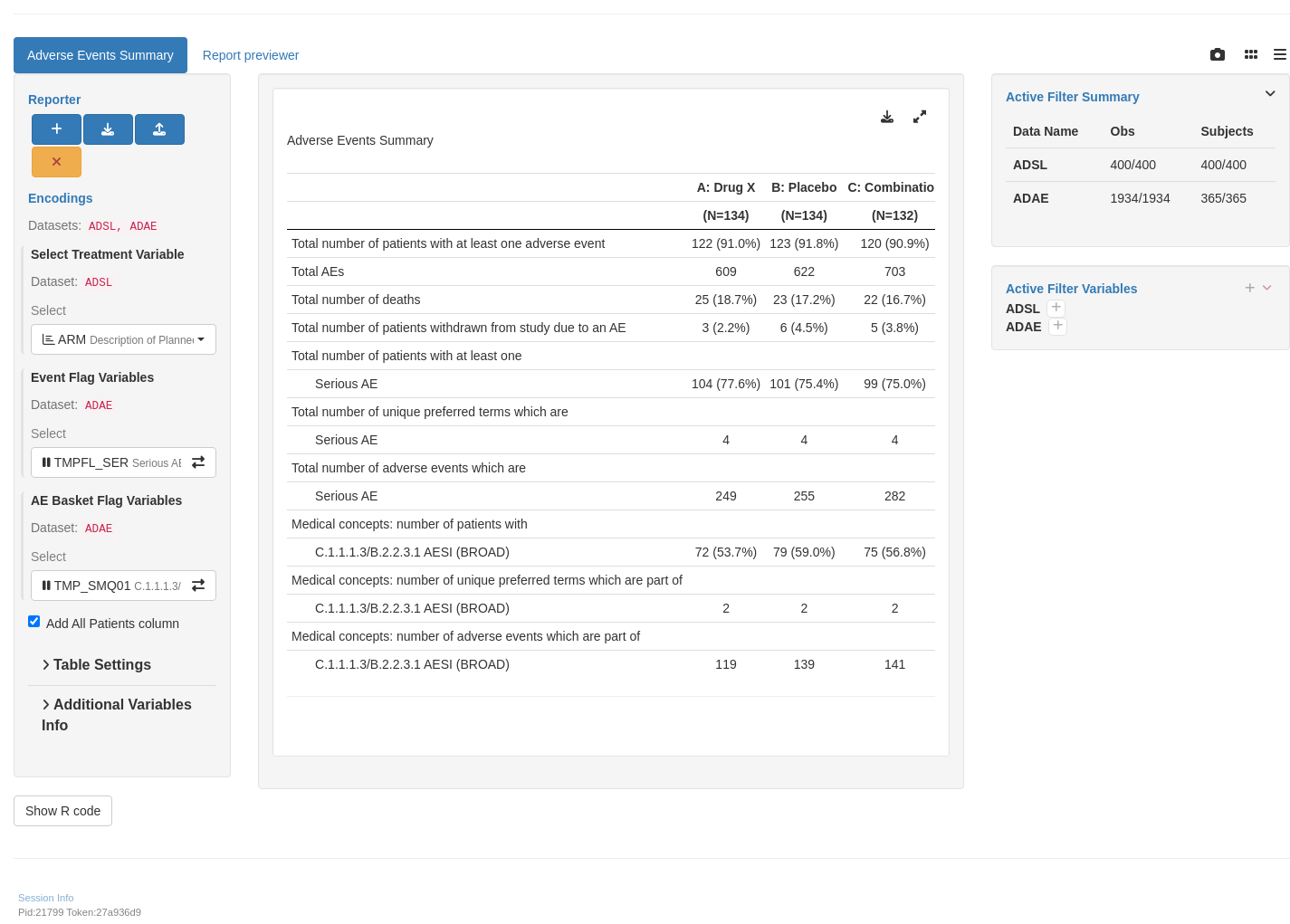
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
library(dplyr)
ADSL <- random.cdisc.data::cadsl
ADAE <- random.cdisc.data::cadae
add_event_flags <- function(dat) {
dat %>%
mutate(
TMPFL_SER = AESER == "Y",
TMPFL_REL = AEREL == "Y",
TMPFL_GR5 = AETOXGR == "5",
TMP_SMQ01 = !is.na(SMQ01NAM),
TMP_SMQ02 = !is.na(SMQ02NAM),
TMP_CQ01 = !is.na(CQ01NAM)
) %>%
col_relabel(
TMPFL_SER = "Serious AE",
TMPFL_REL = "Related AE",
TMPFL_GR5 = "Grade 5 AE",
TMP_SMQ01 = aesi_label(dat[["SMQ01NAM"]], dat[["SMQ01SC"]]),
TMP_SMQ02 = aesi_label(dat[["SMQ02NAM"]], dat[["SMQ02SC"]]),
TMP_CQ01 = aesi_label(dat[["CQ01NAM"]])
)
}
# Generating user-defined event flags.
ADAE <- ADAE %>% add_event_flags()
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADAE")]
## Reusable Configuration For Modules
ADSL <- data[["ADSL"]]
ADAE <- data[["ADAE"]]
ae_anl_vars <- names(ADAE)[startsWith(names(ADAE), "TMPFL_")]
aesi_vars <- names(ADAE)[startsWith(names(ADAE), "TMP_")]
## Setup App
app <- init(
data = data,
modules = modules(
tm_t_events_summary(
label = "Adverse Events Summary",
dataname = "ADAE",
arm_var = choices_selected(
choices = variable_choices("ADSL", c("ARM", "ARMCD")),
selected = "ARM"
),
flag_var_anl = choices_selected(
choices = variable_choices("ADAE", ae_anl_vars),
selected = ae_anl_vars[1],
keep_order = TRUE,
fixed = FALSE
),
flag_var_aesi = choices_selected(
choices = variable_choices("ADAE", aesi_vars),
selected = aesi_vars[1],
keep_order = TRUE,
fixed = FALSE
),
add_total = TRUE
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}