---
title: AET02
subtitle: Adverse Events
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(dplyr)
library(tern)
adsl <- random.cdisc.data::cadsl
adae <- random.cdisc.data::cadae
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
adsl <- df_explicit_na(adsl)
adae <- df_explicit_na(adae) %>%
var_relabel(
AEBODSYS = "MedDRA System Organ Class",
AEDECOD = "MedDRA Preferred Term"
) %>%
filter(ANL01FL == "Y")
# Define the split function
split_fun <- drop_split_levels
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
:::::::::::::::: panel-tabset
## Standard Table
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
add_overall_col(label = "All Patients") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Overall total number of events"
)
) %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Total number of events"
)
) %>%
count_occurrences(
vars = "AEDECOD",
.indent_mods = -1L
) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
result <- build_table(lyt, df = adae, alt_counts_df = adsl)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
The variable `result` corresponds to the adverse events table. However, it includes many empty rows accounting for events which were not reported. The table can be post-processed to prune empty rows and to sort rows, for example by occurrence.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant2, test = list(result_v2 = "result")}
result <- result %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1", "variant2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with <br/> Event Totals
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant3, test = list(result_v3 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
add_overall_col(label = "All Patients") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Overall total number of events"
)
) %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = c("unique"),
.labels = c(
unique = "Total number of patients with at least one adverse event"
)
) %>%
count_occurrences(vars = "AEDECOD", .indent_mods = -1L) %>%
append_varlabels(adae, "AEDECOD", indent = 1L) %>%
count_values(
"STUDYID",
values = as.character(unique(adae$STUDYID)),
.stats = "count",
.labels = c(count = "Total number of events"),
.indent_mods = c(count = -1L)
)
result <- build_table(lyt, df = adae, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences
) %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant3")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with <br/> High-Level Term
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant4, test = list(result_v4 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Overall total number of events"
)
) %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Total number of events"
)
) %>%
split_rows_by(
"AEHLT",
child_labels = "visible",
nested = TRUE,
split_fun = split_fun,
indent_mod = -1L,
label_pos = "topleft",
split_label = obj_label(adae$AEHLT)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Total number of events"
)
) %>%
count_occurrences(vars = "AEDECOD", .indent_mods = -1L) %>%
append_varlabels(adae, c("AEDECOD"), indent = 2L)
result <- build_table(lyt, df = adae, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEHLT"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEHLT", "*", "AEDECOD"),
scorefun = score_occurrences
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant4")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Preferred <br/> Terms Only
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant5, test = list(result_v5 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Total number of events"
),
show_labels = "hidden"
) %>%
count_occurrences(vars = "AEDECOD") %>%
append_varlabels(adae, "AEDECOD")
result <- build_table(lyt, df = adae, alt_counts_df = adsl) %>%
sort_at_path(
path = c("AEDECOD"),
scorefun = score_occurrences
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant5")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Fill-In of <br/> Treatment Groups
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant6, test = list(result_v6 = "result")}
adae_5 <- adae %>% dplyr::filter(ACTARM != "C: Combination")
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
analyze_num_patients(
vars = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Overall total number of events"
)
) %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = c("unique", "nonunique"),
.labels = c(
unique = "Total number of patients with at least one adverse event",
nonunique = "Total number of events"
)
) %>%
count_occurrences(vars = "AEDECOD", .indent_mods = -1L) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
result <- build_table(lyt, df = adae_5, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant6")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table of AEs with an Incidence Rate <br/> $\geq$ 5% in Any Treatment Group (subsetting <br/> preferred terms based on frequency)
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant7, test = list(result_v7 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
indent_mod = -1L,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
count_occurrences(
vars = "AEDECOD",
.indent_mods = 1L
) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
result <- build_table(lyt, df = adae, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences
)
criteria_fun <- function(tr) is(tr, "ContentRow")
result <- trim_rows(result, criteria = criteria_fun)
row_condition <- has_fraction_in_any_col(
atleast = 0.05,
col_names = names(table(adsl$ACTARM))
)
result <- prune_table(result, keep_rows(row_condition))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant7")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table of AEs with an Incidence Rate $\geq$ 5% in <br/> Any Treatment Group (subsetting preferred terms <br/> based on frequency with high-level terms)
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant8, test = list(result_v8 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = TRUE,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
split_rows_by(
"AEHLT",
child_labels = "visible",
nested = TRUE,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEHLT)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
count_occurrences(vars = "AEDECOD") %>%
append_varlabels(adae, c("AEDECOD"), indent = 2L)
result <- build_table(lyt, adae, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols,
decreasing = TRUE
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEHLT"),
scorefun = cont_n_allcols,
decreasing = TRUE
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEHLT", "*", "AEDECOD"),
scorefun = score_occurrences,
decreasing = TRUE
)
criteria_fun <- function(tr) is(tr, "ContentRow")
result <- trim_rows(result, criteria = criteria_fun)
row_condition <- has_fraction_in_any_col(
atleast = 0.05,
col_names = names(table(adsl$ACTARM))
)
result <- prune_table(result, keep_rows(row_condition))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant8")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table of AEs with an Incidence Rate $\geq$ 10% <br/> in Any Treatment Group (subsetting <br/> preferred terms based on frequency)
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant9, test = list(result_v9 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
indent_mod = -1L,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
count_occurrences(
vars = "AEDECOD",
.indent_mods = 1L
) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
result <- build_table(lyt, df = adae, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences
)
criteria_fun <- function(tr) is(tr, "ContentRow")
result <- trim_rows(result, criteria = criteria_fun)
row_condition <- has_fraction_in_any_col(
atleast = 0.10,
col_names = names(table(adsl$ACTARM))
)
result <- prune_table(result, keep_rows(row_condition))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant9")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table of AEs with an Incidence Rate $\geq$ 3 <br/> Patients in Any Treatment Group (subsetting <br/> preferred terms based on number of patients)
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant10, test = list(result_v10 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
indent_mod = -1L,
split_fun = split_fun,
split_label = obj_label(adae$AEBODSYS),
label_pos = "topleft"
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
count_occurrences(
vars = "AEDECOD",
.indent_mods = 1L
) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
result <- build_table(lyt, df = adae, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences
)
criteria_fun <- function(tr) is(tr, "ContentRow")
result <- trim_rows(result, criteria = criteria_fun)
row_condition <- has_count_in_any_col(
atleast = 3,
col_names = names(table(adsl$ACTARM))
)
result <- prune_table(result, keep_rows(row_condition))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant10")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table of AEs with a Difference in Incidence Rate $\geq$ 5% <br/> Between Any Treatment (subsetting preferred terms based <br/> on difference in percentage between treatment groups)
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant11, test = list(result_v11 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
indent_mod = -1L,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
count_occurrences(
vars = "AEDECOD",
.indent_mods = 1L
) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
result <- build_table(lyt, df = adae, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences
)
criteria_fun <- function(tr) is(tr, "ContentRow")
result <- trim_rows(result, criteria = criteria_fun)
row_condition <- has_fractions_difference(
atleast = 0.05,
col_names = levels(adsl$ACTARM)
)
result <- prune_table(result, keep_rows(row_condition))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant11")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table of AEs with an Incidence Rate $\geq$ 5% <br/> in B: Placebo (subsetting preferred terms based <br/> on frequency for a particular treatment group)
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant12, test = list(result_v12 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
indent_mod = -1L,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
count_occurrences(
vars = "AEDECOD",
.indent_mods = 1L
) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
result <- build_table(lyt, df = adae, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences
)
criteria_fun <- function(tr) is(tr, "ContentRow")
result <- trim_rows(result, criteria = criteria_fun)
row_condition <- has_fraction_in_cols(
atleast = 0.05,
col_names = c("B: Placebo")
)
result <- prune_table(result, keep_rows(row_condition))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant12")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table of AEs with a Difference in Incidence Rate $\geq$ 5% Between <br/> Arm A and Arm B or Arm C (displaying preferred terms with a <br/> difference of at least x% between selected treatment groups)
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant13, test = list(result_v13 = "result")}
lyt <- basic_table(show_colcounts = TRUE) %>%
split_cols_by(var = "ACTARM") %>%
split_rows_by(
"AEBODSYS",
child_labels = "visible",
nested = FALSE,
indent_mod = -1L,
split_fun = split_fun,
label_pos = "topleft",
split_label = obj_label(adae$AEBODSYS)
) %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
count_occurrences(
vars = "AEDECOD",
.indent_mods = 1L
) %>%
append_varlabels(adae, "AEDECOD", indent = 1L)
result <- build_table(lyt, df = adae, alt_counts_df = adsl) %>%
prune_table() %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = cont_n_allcols
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences
)
criteria_fun <- function(tr) is(tr, "ContentRow")
result <- trim_rows(result, criteria = criteria_fun)
row_condition1 <- has_fractions_difference(atleast = 0.05, col_names = c("A: Drug X", "B: Placebo"))
row_condition2 <- has_fractions_difference(atleast = 0.05, col_names = c("A: Drug X", "C: Combination"))
row_condition <- row_condition1 | row_condition2
result <- prune_table(result, keep_rows(row_condition))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant13")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
::::::::::::::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
ADSL <- random.cdisc.data::cadsl
ADAE <- random.cdisc.data::cadae
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
ADSL <- df_explicit_na(ADSL)
ADAE <- df_explicit_na(ADAE)
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADAE")]
## Reusable Configuration For Modules
ADAE <- data[["ADAE"]]
## Setup App
app <- init(
data = data,
modules = modules(
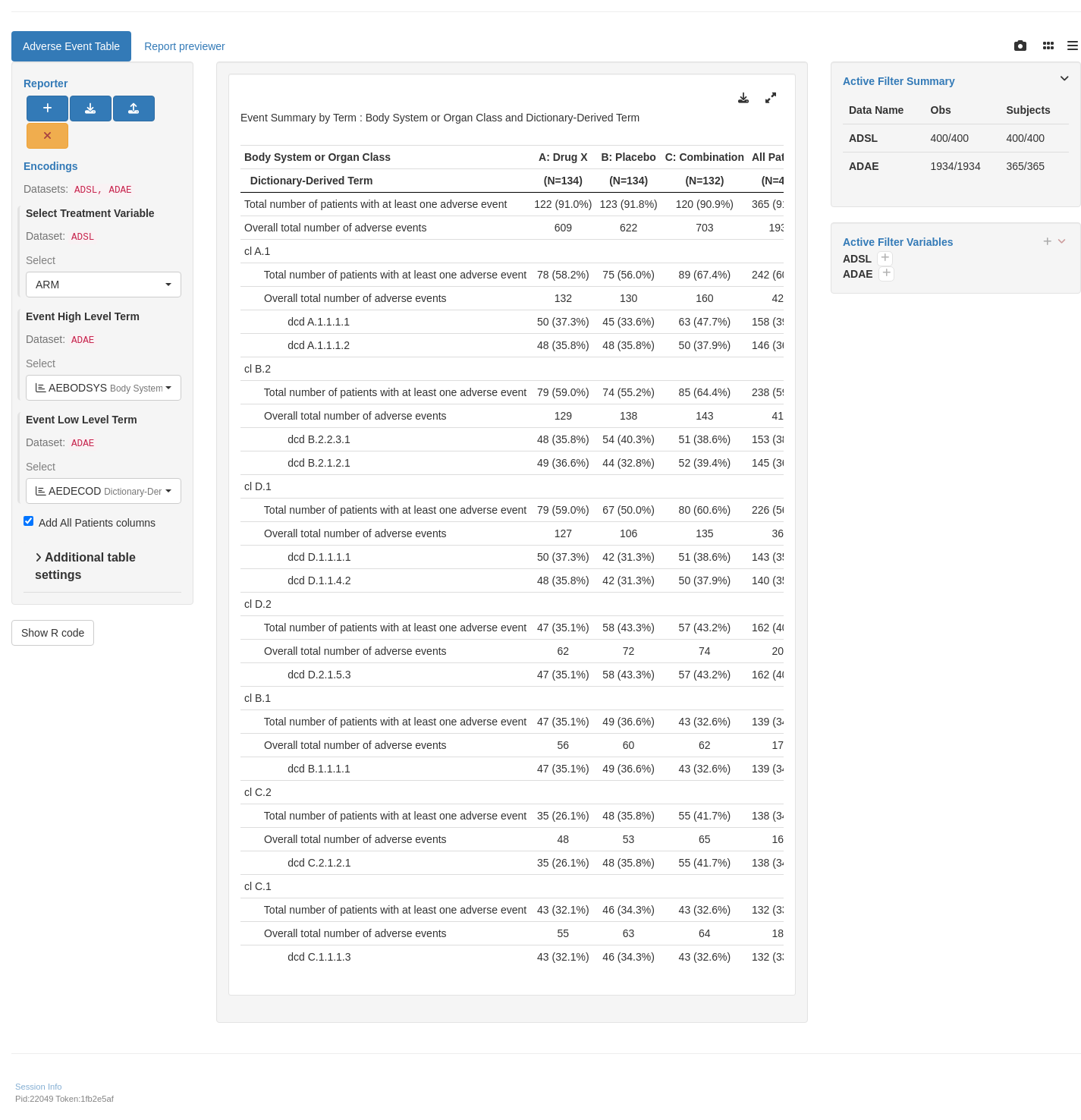
tm_t_events(
label = "Adverse Event Table",
dataname = "ADAE",
arm_var = choices_selected(c("ARM", "ARMCD"), "ARM"),
llt = choices_selected(
choices = variable_choices(ADAE, c("AETERM", "AEDECOD")),
selected = c("AEDECOD")
),
hlt = choices_selected(
choices = variable_choices(ADAE, c("AEBODSYS", "AESOC")),
selected = "AEBODSYS"
),
add_total = TRUE,
event_type = "adverse event"
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}