---
title: AET04_PI
subtitle: Adverse Events Reported in $\geq$ 10% of Patients by Highest NCI CTCAE Grade
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(dplyr)
library(tern)
preprocess_adae <- function(adae) {
adae %>%
dplyr::group_by(ACTARM, USUBJID, AEBODSYS, AEDECOD) %>%
dplyr::summarize(
MAXAETOXGR = max(as.numeric(AETOXGR)),
.groups = "drop"
) %>%
dplyr::ungroup() %>%
dplyr::mutate(
MAXAETOXGR = factor(MAXAETOXGR),
AEDECOD = droplevels(as.factor(AEDECOD))
)
}
full_table_aet04_pi <- function(adsl, adae_max) {
grade_groups <- list(
"Any Grade (%)" = c("1", "2", "3", "4", "5"),
"Grade 3-4 (%)" = c("3", "4"),
"Grade 5 (%)" = "5"
)
col_counts <- rep(table(adsl$ACTARM), each = length(grade_groups))
basic_table() %>%
split_cols_by("ACTARM") %>%
split_cols_by_groups("MAXAETOXGR", groups_list = grade_groups) %>%
split_rows_by("AEBODSYS",
child_labels = "visible", nested = FALSE, indent_mod = -1L,
split_fun = trim_levels_in_group("AEDECOD")
) %>%
append_topleft("MedDRA System Organ Class") %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
analyze_vars(
"AEDECOD",
na.rm = FALSE,
denom = "N_col",
.stats = "count_fraction",
.formats = c(count_fraction = format_fraction_threshold(0.01))
) %>%
append_topleft(" MedDRA Preferred Term") %>%
build_table(adae_max, col_counts = col_counts)
}
criteria_fun <- function(tr) {
is(tr, "ContentRow")
}
adsl <- random.cdisc.data::cadsl
adae_max <- random.cdisc.data::cadae %>%
preprocess_adae() %>%
df_explicit_na()
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
::::::::::: panel-tabset
## Standard Table
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant1, test = list(result_v1 = "result")}
full_table <- full_table_aet04_pi(adsl, adae_max) %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = score_occurrences_cont_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
)
at_least_10percent_any <- has_fraction_in_any_col(atleast = 0.1, col_indices = c(1, 4, 7))
result <- full_table %>%
trim_rows(criteria = criteria_fun) %>%
prune_table(keep_rows(at_least_10percent_any))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Table with Patients Treated with a Particular Treatment by Highest NCI <br/> CTCAE Grade (specifying a treatment for selecting preferred terms)
Note: User needs to specify the column index for filtering the table. The current example uses the "Any Grade" column for treatment A: Drug X with a filtering threshold at 0.37; AEs reported in greater than 37% of patients in treatment A: Drug X are shown. This can be changed by varying the parameter values in the `has_fraction_in_cols` function.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant2, test = list(result_v2 = "result")}
full_table <- full_table_aet04_pi(adsl, adae_max) %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = score_occurrences_cont_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
)
at_least_37percent_any_drugx <- has_fraction_in_cols(atleast = 0.37, col_indices = 1)
result <- full_table %>%
trim_rows(criteria = criteria_fun) %>%
prune_table(keep_rows(at_least_37percent_any_drugx))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Adverse Events Reported in $\geq$ 5% of Patients by <br/> Highest NCI CTCAE Grade (changing the threshold)
Note: User needs to specify the column index for filtering the table. The current example uses column indices 1, 4, and 7 with a filtering threshold at 0.40 to demonstrate the filtering ability; AEs reported in greater than 40% of patients are shown. This can be changed by varying the parameter values in the `has_fraction_in_any_col` function.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant3, test = list(result_v3 = "result")}
full_table <- full_table_aet04_pi(adsl, adae_max) %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = score_occurrences_cont_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
)
at_least_40percent_any <- has_fraction_in_any_col(atleast = 0.40, col_indices = c(1, 4, 7))
result <- full_table %>%
trim_rows(criteria = criteria_fun) %>%
prune_table(keep_rows(at_least_40percent_any))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant3")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Adverse Events Reported in $\geq$ 5% of Patients and <br/> $\geq$ 2% Difference Between Treatments by Highest <br/> NCI CTCAE Grade (using more than one condition)
Note: User needs to specify the column index for filtering the table. The current example uses column indices 1, 4, and 7 to filter for AEs reported in $\geq$ 30% of patients and AEs reported with a $\geq$ 15% difference between treatments to demonstrate the filtering ability; this can be changed by varying the parameter values in the `has_fraction_in_any_col` and `has_fractions_difference` functions.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant4, test = list(result_v4 = "result")}
full_table <- full_table_aet04_pi(adsl, adae_max) %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = score_occurrences_cont_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
)
at_least_30percent_any <- has_fraction_in_any_col(atleast = 0.3, col_indices = c(1, 4, 7))
at_least_15percent_diff <- has_fractions_difference(atleast = 0.15, col_indices = c(1, 4, 7))
result <- full_table %>%
trim_rows(criteria = criteria_fun) %>%
prune_table(keep_rows(at_least_30percent_any & at_least_15percent_diff))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant4")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Adverse Events Reported in $\geq$ 10% of Patients for Any Grade <br/> or $\geq$ 5% of Patients for Grade 3-4 by Highest NCI CTCAE <br/> Grade (using different condition depending on the grade)
Note: User needs to specify the column index for filtering the table. The current example filters using column indices 1, 4, and 7 to include AEs reported in $\geq$ 40% of patients and using column indices 2, 5, and 8 to include grade 3-4 AEs reported in $\geq$ 20% of patients. These thresholds are chosen to demonstrate the filtering ability; they can be changed by varying parameter values in the `has_fraction_in_any_col` function.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant5, test = list(result_v5 = "result")}
full_table <- full_table_aet04_pi(adsl, adae_max) %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = score_occurrences_cont_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
)
at_least_40percent_any <- has_fraction_in_any_col(atleast = 0.4, col_indices = c(1, 4, 7))
at_least_20percent_g34 <- has_fraction_in_any_col(atleast = 0.20, col_indices = c(2, 5, 8))
result <- full_table %>%
trim_rows(criteria = criteria_fun) %>%
prune_table(keep_rows(at_least_40percent_any | at_least_20percent_g34))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant5")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Standard Table with <br/> Modified Grade Grouping
Note: User can specify/modify the grouping of grades, as shown in this example. In addition, the user needs to specify the column index for filtering the table. The current example uses the "Any Grade" columns with a filtering threshold at 0.40; this can be changed by varying the parameter values in the `has_fraction_in_any_col` function.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant6, test = list(result_v6 = "result")}
grade_groups <- list(
"Any Grade (%)" = c("1", "2", "3", "4", "5"),
"Grade 1-2 (%)" = c("1", "2"),
"Grade 3-4 (%)" = c("3", "4"),
"Grade 5 (%)" = "5"
)
col_counts <- rep(table(adsl$ACTARM), each = length(grade_groups))
full_table <- basic_table() %>%
split_cols_by("ACTARM") %>%
split_cols_by_groups("MAXAETOXGR", groups_list = grade_groups) %>%
split_rows_by("AEBODSYS",
child_labels = "visible", nested = FALSE, indent_mod = -1L,
split_fun = trim_levels_in_group("AEDECOD")
) %>%
append_topleft("MedDRA System Organ Class") %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
analyze_vars(
"AEDECOD",
na.rm = FALSE,
denom = "N_col",
.stats = "count_fraction",
.formats = c(count_fraction = format_fraction_threshold(0.01))
) %>%
append_topleft(" MedDRA Preferred Term") %>%
build_table(adae_max, col_counts = col_counts) %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = score_occurrences_cont_cols(col_indices = c(1, 5, 9)),
decreasing = TRUE
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences_cols(col_indices = c(1, 5, 9)),
decreasing = TRUE
)
at_least_40percent_any <- has_fraction_in_any_col(atleast = 0.4, col_indices = c(1, 5, 9))
result <- full_table %>%
trim_rows(criteria = criteria_fun) %>%
prune_table(keep_rows(at_least_40percent_any))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant6")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Standard Table with <br/> Overlapping Grade Groupings
Note: User needs to specify the column index for filtering the table. The current example uses column indices 1, 5, and 9 with a filtering threshold at 0.40 to demonstrate the filtering ability; any grade AEs reported in greater than 40% of patients are shown. This can be changed by varying the parameter values in the `has_fraction_in_any_col` function.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant7, test = list(result_v7 = "result")}
grade_groups <- list(
"Any Grade (%)" = c("1", "2", "3", "4", "5"),
"Grade 3-4 (%)" = c("3", "4"),
"Grade 3-5 (%)" = c("3", "4", "5"),
"Grade 5 (%)" = "5"
)
col_counts <- rep(table(adsl$ACTARM), each = length(grade_groups))
full_table <- basic_table() %>%
split_cols_by("ACTARM") %>%
split_cols_by_groups("MAXAETOXGR", groups_list = grade_groups) %>%
split_rows_by("AEBODSYS",
child_labels = "visible", nested = FALSE, indent_mod = -1L,
split_fun = trim_levels_in_group("AEDECOD")
) %>%
append_topleft("MedDRA System Organ Class") %>%
summarize_num_patients(
var = "USUBJID",
.stats = "unique",
.labels = "Total number of patients with at least one adverse event"
) %>%
analyze_vars(
"AEDECOD",
na.rm = FALSE,
denom = "N_col",
.stats = "count_fraction",
.formats = c(count_fraction = format_fraction_threshold(0.01))
) %>%
append_topleft(" MedDRA Preferred Term") %>%
build_table(adae_max, col_counts = col_counts) %>%
sort_at_path(
path = c("AEBODSYS"),
scorefun = score_occurrences_cont_cols(col_indices = c(1, 5, 9)),
decreasing = TRUE
) %>%
sort_at_path(
path = c("AEBODSYS", "*", "AEDECOD"),
scorefun = score_occurrences_cols(col_indices = c(1, 5, 9)),
decreasing = TRUE
)
at_least_40percent_any <- has_fraction_in_any_col(atleast = 0.4, col_indices = c(1, 5, 9))
result <- full_table %>%
trim_rows(criteria = criteria_fun) %>%
prune_table(keep_rows(at_least_40percent_any))
result
```
```{r include = FALSE}
webr_code_labels <- c("variant7")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Standard Table without SOCs
Note: User needs to specify the column index for filtering the table. The current example uses column indices 1, 4, and 7 with a filtering threshold at 0.20 to demonstrate the filtering ability; any grade AEs (Preferred Terms Only) reported in greater than 20% of patients are shown. This can be changed by varying the parameter values in the `has_fraction_in_any_col` function.
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r variant8, test = list(result_v8 = "result")}
grade_groups <- list(
"Any Grade (%)" = c("1", "2", "3", "4", "5"),
"Grade 3-4 (%)" = c("3", "4"),
"Grade 5 (%)" = "5"
)
col_counts <- rep(table(adsl$ACTARM), each = length(grade_groups))
full_table <- basic_table() %>%
split_cols_by("ACTARM") %>%
split_cols_by_groups("MAXAETOXGR", groups_list = grade_groups) %>%
analyze_vars(
"AEDECOD",
na.rm = FALSE,
denom = "N_col",
.stats = "count_fraction",
.formats = c(count_fraction = format_fraction_threshold(0.01))
) %>%
append_topleft("MedDRA Preferred Term") %>%
build_table(adae_max, col_counts = col_counts)
at_least_20percent_any <- has_fraction_in_any_col(atleast = 0.2, col_indices = c(1, 4, 7))
result <- full_table %>%
prune_table(keep_rows(at_least_20percent_any)) %>%
sort_at_path(
path = c("AEDECOD"),
scorefun = score_occurrences_cols(col_indices = c(1, 4, 7)),
decreasing = TRUE
)
result
```
```{r include = FALSE}
webr_code_labels <- c("variant8")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
:::::::::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
ADSL <- random.cdisc.data::cadsl
ADAE <- random.cdisc.data::cadae
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADAE")]
## Reusable Configuration For Modules
ADAE <- data[["ADAE"]]
## Setup App
app <- init(
data = data,
modules = modules(
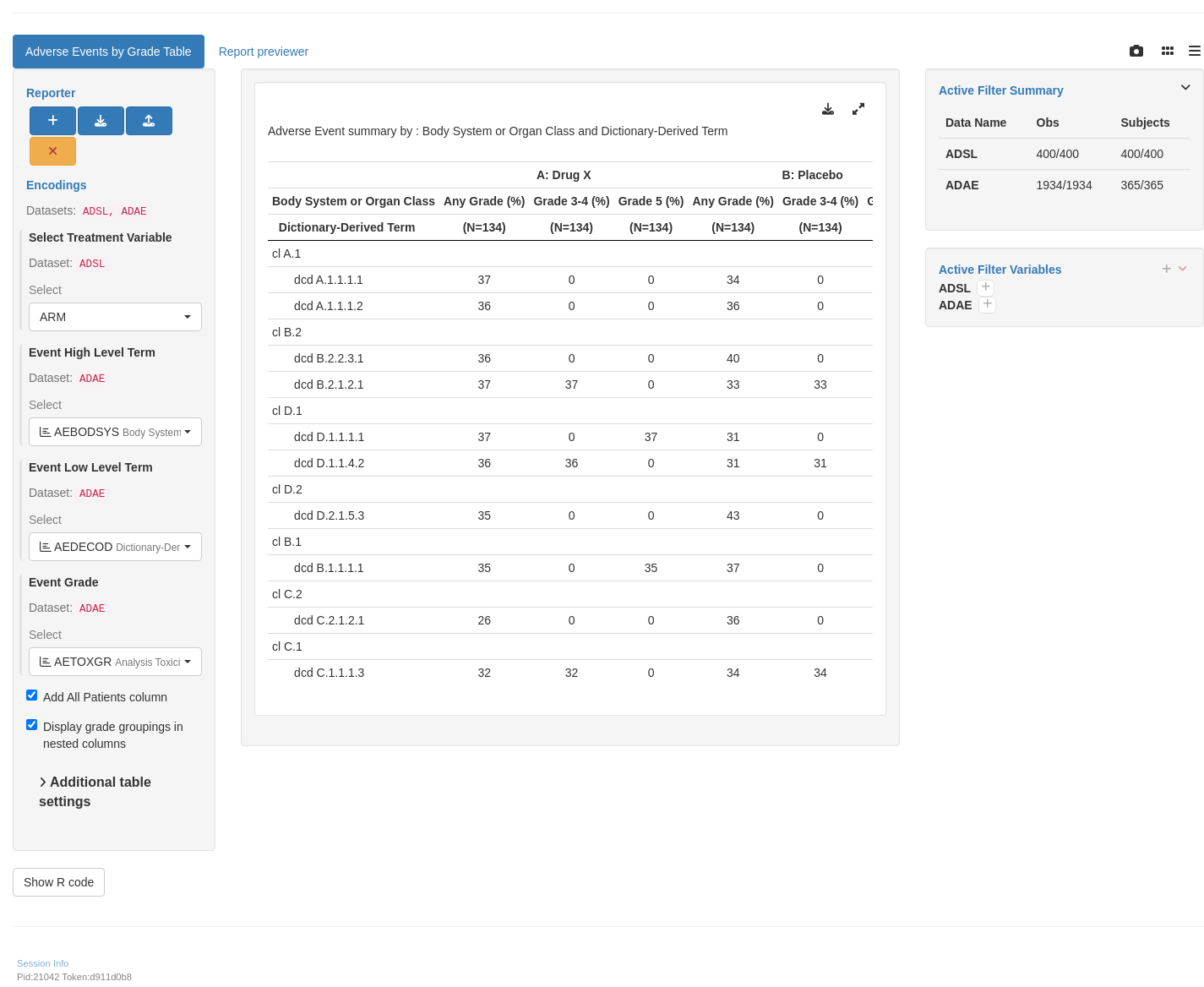
tm_t_events_by_grade(
label = "Adverse Events by Grade Table",
dataname = "ADAE",
arm_var = choices_selected(c("ARM", "ARMCD"), "ARM"),
col_by_grade = TRUE,
llt = choices_selected(
choices = variable_choices(ADAE, c("AETERM", "AEDECOD")),
selected = c("AEDECOD")
),
hlt = choices_selected(
choices = variable_choices(ADAE, c("AEBODSYS", "AESOC")),
selected = "AEBODSYS"
),
grade = choices_selected(
choices = variable_choices(ADAE, c("AETOXGR", "AESEV")),
selected = "AETOXGR"
),
# List of column groups for grades used when `col_by_grade` = TRUE.
grading_groups = list(
`Any Grade (%)` = c("1", "2", "3", "4", "5"),
`Grade 3-4 (%)` = c("3", "4"),
`Grade 5 (%)` = "5"
)
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}