Code
[[1]]
[[2]]Individual Patient Plot Over Time
For illustration purposes, we will subset the adlb dataset for safety population in treatment arm A and a specific lab parameter (ALT).
The user can select different plotting_choices depending on their preference. To demonstrate, separate plots are produced with a maximum of 3 observations each.
Here, patients’ individual baseline values will be shown for reference. Note that users can provide their own custom theme to the function via the ggtheme argument.
plots <- g_ipp(
df = adlb_f,
xvar = "AVISIT",
yvar = "AVAL",
xlab = "Visit",
ylab = "SGOT/ALT (U/L)",
id_var = "Patient_ID",
title = "Individual Patient Plots",
subtitle = "Treatment Arm A",
add_baseline_hline = TRUE,
yvar_baseline = "BASE",
ggtheme = theme_minimal(),
plotting_choices = "split_by_max_obs",
max_obs_per_plot = 3
)
plots[[1]]
[[2]]library(tern)
library(dplyr)
library(ggplot2)
library(nestcolor)
# use small sample size
adsl <- random.cdisc.data::cadsl %>% slice(1:15)
adlb <- random.cdisc.data::cadlb %>% filter(USUBJID %in% adsl$USUBJID)
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
adlb <- df_explicit_na(adlb)
adlb_f <- adlb %>%
filter(
SAFFL == "Y",
PARAMCD == "ALT",
AVISIT != "SCREENING",
ARMCD == "ARM A"
) %>%
mutate(Patient_ID = sub(".*id-", "", USUBJID))teal Applibrary(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
library(dplyr)
# use small sample size
ADSL <- random.cdisc.data::cadsl %>% slice(1:15)
ADLB <- random.cdisc.data::cadlb %>% filter(USUBJID %in% ADSL$USUBJID)
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
ADSL <- df_explicit_na(ADSL)
ADLB <- df_explicit_na(ADLB) %>%
filter(AVISIT != "SCREENING")
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADLB")]
## Reusable Configuration For Modules
ADLB <- data[["ADLB"]]
## Setup App
app <- init(
data = data,
modules = modules(
tm_g_ipp(
label = "Individual Patient Plot",
dataname = "ADLB",
arm_var = choices_selected(
value_choices(ADLB, c("ARMCD")),
"ARM A"
),
paramcd = choices_selected(
value_choices(ADLB, "PARAMCD"),
"ALT"
),
aval_var = choices_selected(
variable_choices(ADLB, c("AVAL")),
"AVAL"
),
avalu_var = choices_selected(
variable_choices(ADLB, c("AVALU")),
"AVALU",
fixed = TRUE
),
id_var = choices_selected(
variable_choices(ADLB, c("USUBJID")),
"USUBJID",
fixed = TRUE
),
visit_var = choices_selected(
variable_choices(ADLB, c("AVISIT")),
"AVISIT"
),
baseline_var = choices_selected(
variable_choices(ADLB, c("BASE")),
"BASE",
fixed = TRUE
),
add_baseline_hline = FALSE,
separate_by_obs = FALSE
)
)
)
shinyApp(app$ui, app$server)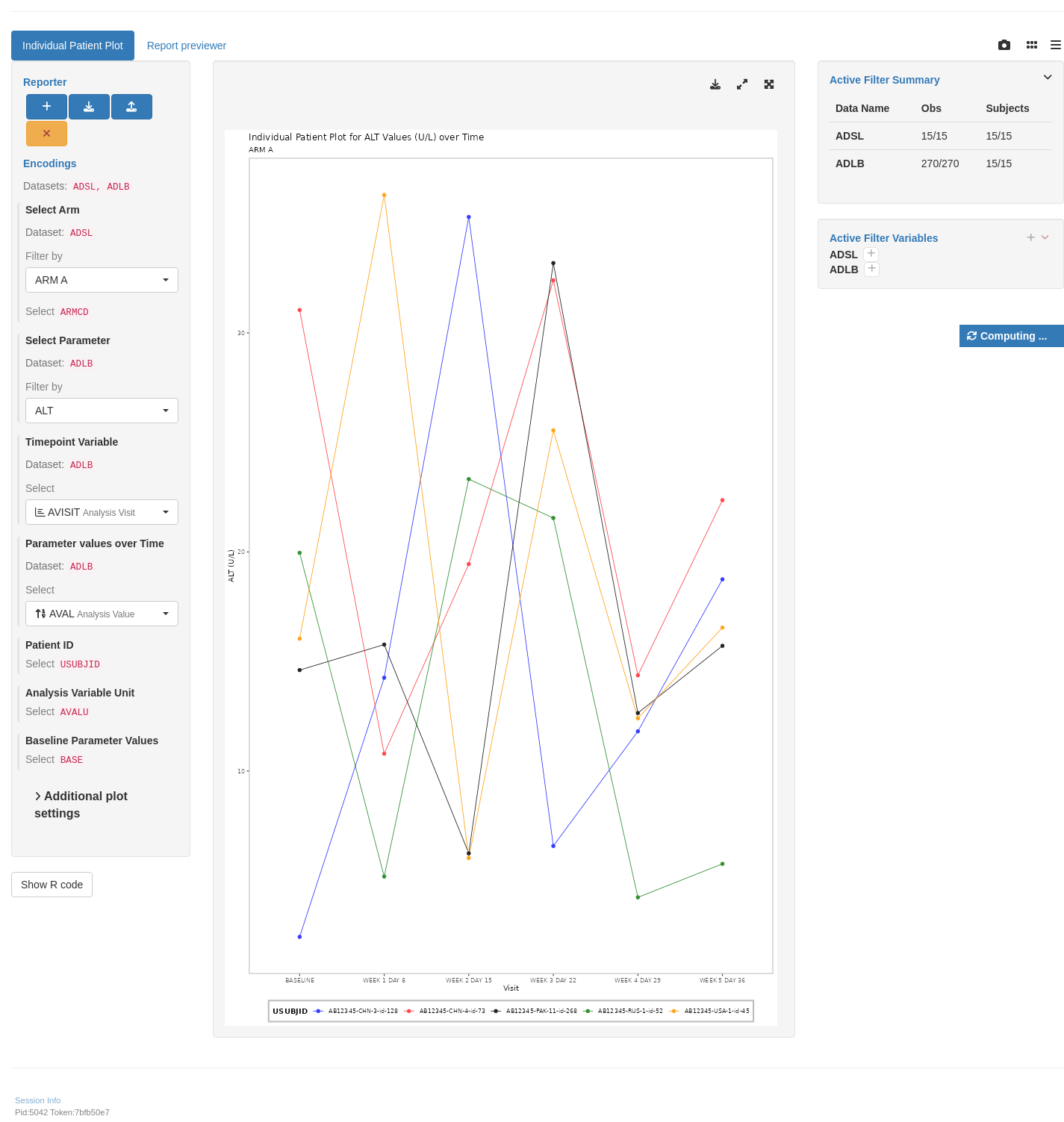
shinylive allow you to modify to run shiny application entirely in the web browser. Modify the code below and click re-run the app to see the results. The performance is slighly worse and some of the features (e.g. downloading) might not work at all.
#| '!! shinylive warning !!': |
#| shinylive does not work in self-contained HTML documents.
#| Please set `embed-resources: false` in your metadata.
#| standalone: true
#| viewerHeight: 800
#| editorHeight: 200
#| components: [viewer, editor]
#| layout: vertical
# -- WEBR HELPERS --
options(webr_pkg_repos = c("r-universe" = "https://pharmaverse.r-universe.dev", getOption("webr_pkg_repos")))
# -- APP CODE --
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
library(dplyr)
# use small sample size
ADSL <- random.cdisc.data::cadsl %>% slice(1:15)
ADLB <- random.cdisc.data::cadlb %>% filter(USUBJID %in% ADSL$USUBJID)
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
ADSL <- df_explicit_na(ADSL)
ADLB <- df_explicit_na(ADLB) %>%
filter(AVISIT != "SCREENING")
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADLB")]
## Reusable Configuration For Modules
ADLB <- data[["ADLB"]]
## Setup App
app <- init(
data = data,
modules = modules(
tm_g_ipp(
label = "Individual Patient Plot",
dataname = "ADLB",
arm_var = choices_selected(
value_choices(ADLB, c("ARMCD")),
"ARM A"
),
paramcd = choices_selected(
value_choices(ADLB, "PARAMCD"),
"ALT"
),
aval_var = choices_selected(
variable_choices(ADLB, c("AVAL")),
"AVAL"
),
avalu_var = choices_selected(
variable_choices(ADLB, c("AVALU")),
"AVALU",
fixed = TRUE
),
id_var = choices_selected(
variable_choices(ADLB, c("USUBJID")),
"USUBJID",
fixed = TRUE
),
visit_var = choices_selected(
variable_choices(ADLB, c("AVISIT")),
"AVISIT"
),
baseline_var = choices_selected(
variable_choices(ADLB, c("BASE")),
"BASE",
fixed = TRUE
),
add_baseline_hline = FALSE,
separate_by_obs = FALSE
)
)
)
shinyApp(app$ui, app$server)[1] "2026-03-07 17:33:21 UTC"─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.4 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Etc/UTC
date 2026-03-07
pandoc 3.9 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
backports 1.5.0 2024-05-23 [1] RSPM
brio 1.1.5 2024-04-24 [1] RSPM
broom 1.0.12 2026-01-27 [1] RSPM
bsicons 0.1.2 2023-11-04 [1] RSPM
bslib 0.10.0 2026-01-26 [1] RSPM
cachem 1.1.0 2024-05-16 [1] RSPM
callr 3.7.6 2024-03-25 [1] RSPM
checkmate 2.3.4 2026-02-03 [1] RSPM
chromote 0.5.1 2025-04-24 [1] RSPM
cli 3.6.5 2025-04-23 [1] RSPM
codetools 0.2-20 2024-03-31 [2] CRAN (R 4.5.2)
curl 7.0.0 2025-08-19 [1] RSPM
dichromat 2.0-0.1 2022-05-02 [1] CRAN (R 4.5.2)
digest 0.6.39 2025-11-19 [1] RSPM
dplyr * 1.2.0 2026-02-03 [1] RSPM
evaluate 1.0.5 2025-08-27 [1] RSPM
farver 2.1.2 2024-05-13 [1] RSPM
fastmap 1.2.0 2024-05-15 [1] RSPM
fontawesome 0.5.3 2024-11-16 [1] RSPM
forcats 1.0.1 2025-09-25 [1] RSPM
formatR 1.14 2023-01-17 [1] CRAN (R 4.5.2)
formatters * 0.5.12.9002 2026-01-03 [1] https://p~
fs 1.6.7 2026-03-06 [1] RSPM
generics 0.1.4 2025-05-09 [1] RSPM
ggplot2 * 4.0.2 2026-02-03 [1] RSPM
glue 1.8.0 2024-09-30 [1] RSPM
gtable 0.3.6 2024-10-25 [1] RSPM
htmltools 0.5.9 2025-12-04 [1] RSPM
htmlwidgets 1.6.4 2023-12-06 [1] RSPM
httpuv 1.6.16 2025-04-16 [1] RSPM
jquerylib 0.1.4 2021-04-26 [1] RSPM
jsonlite 2.0.0 2025-03-27 [1] RSPM
knitr 1.51 2025-12-20 [1] RSPM
labeling 0.4.3 2023-08-29 [1] RSPM
later 1.4.8 2026-03-05 [1] RSPM
lattice 0.22-9 2026-02-09 [2] CRAN (R 4.5.2)
lifecycle 1.0.5 2026-01-08 [1] RSPM
logger 0.4.1 2025-09-11 [1] RSPM
magrittr * 2.0.4 2025-09-12 [1] RSPM
Matrix 1.7-4 2025-08-28 [1] CRAN (R 4.5.2)
memoise 2.0.1 2021-11-26 [1] RSPM
mime 0.13 2025-03-17 [1] RSPM
nestcolor * 0.1.3.9000 2025-01-21 [1] https://p~
otel 0.2.0 2025-08-29 [1] RSPM
pillar 1.11.1 2025-09-17 [1] RSPM
pkgcache 2.2.4 2025-05-26 [1] RSPM
pkgconfig 2.0.3 2019-09-22 [1] RSPM
processx 3.8.6 2025-02-21 [1] RSPM
promises 1.5.0 2025-11-01 [1] RSPM
ps 1.9.1 2025-04-12 [1] RSPM
purrr 1.2.1 2026-01-09 [1] RSPM
R6 2.6.1 2025-02-15 [1] RSPM
ragg 1.5.1 2026-03-06 [1] RSPM
random.cdisc.data 0.3.16.9007 2025-11-13 [1] https://p~
rbibutils 2.4.1 2026-01-21 [1] RSPM
RColorBrewer 1.1-3 2022-04-03 [1] RSPM
Rcpp 1.1.1 2026-01-10 [1] RSPM
Rdpack 2.6.6 2026-02-08 [1] RSPM
rlang 1.1.7 2026-01-09 [1] RSPM
rmarkdown 2.30 2025-09-28 [1] RSPM
rtables * 0.6.15.9003 2026-01-03 [1] https://p~
S7 0.2.1 2025-11-14 [1] RSPM
sass 0.4.10 2025-04-11 [1] RSPM
scales 1.4.0 2025-04-24 [1] RSPM
sessioninfo 1.2.3 2025-02-05 [1] any (@1.2.3)
shiny * 1.13.0 2026-02-20 [1] RSPM
shinycssloaders 1.1.0 2024-07-30 [1] RSPM
shinyjs 2.1.1 2026-01-15 [1] RSPM
shinyvalidate 0.1.3 2023-10-04 [1] RSPM
shinyWidgets 0.9.0 2025-02-21 [1] RSPM
stringi 1.8.7 2025-03-27 [1] RSPM
stringr 1.6.0 2025-11-04 [1] RSPM
survival 3.8-6 2026-01-16 [2] CRAN (R 4.5.2)
systemfonts 1.3.2 2026-03-05 [1] RSPM
teal * 1.1.0.9029 2026-03-06 [1] https://p~
teal.code * 0.7.1.9000 2026-01-21 [1] https://p~
teal.data * 0.8.0.9002 2025-12-15 [1] https://p~
teal.logger 0.4.1.9004 2026-02-10 [1] https://p~
teal.modules.clinical * 0.12.0.9011 2026-03-02 [1] https://p~
teal.reporter 0.6.1.9000 2026-02-20 [1] https://p~
teal.slice * 0.7.1.9006 2026-02-25 [1] https://p~
teal.transform * 0.7.1.9003 2026-02-17 [1] https://p~
teal.widgets 0.6.0.9000 2026-02-24 [1] https://p~
tern * 0.9.10.9006 2026-01-03 [1] https://p~
testthat 3.3.2 2026-01-11 [1] RSPM
textshaping 1.0.5 2026-03-06 [1] RSPM
tibble 3.3.1 2026-01-11 [1] RSPM
tidyr 1.3.2 2025-12-19 [1] RSPM
tidyselect 1.2.1 2024-03-11 [1] RSPM
vctrs 0.7.1 2026-01-23 [1] RSPM
webshot 0.5.5 2023-06-26 [1] CRAN (R 4.5.2)
webshot2 0.1.2 2025-04-23 [1] RSPM
websocket 1.4.4 2025-04-10 [1] RSPM
withr 3.0.2 2024-10-28 [1] RSPM
xfun 0.56 2026-01-18 [1] RSPM
xtable 1.8-8 2026-02-22 [1] RSPM
yaml 2.3.12 2025-12-10 [1] RSPM
[1] /usr/local/lib/R/site-library
[2] /usr/local/lib/R/library
[3] /github/home/R/x86_64-pc-linux-gnu-library/4.5
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────.lock fileDownload the .lock file and use renv::restore() on it to recreate environment used to generate this website.
---
title: IPPG01
subtitle: Individual Patient Plot Over Time
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
For illustration purposes, we will subset the `adlb` dataset for safety population in treatment arm A and a specific lab parameter (`ALT`).
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(tern)
library(dplyr)
library(ggplot2)
library(nestcolor)
# use small sample size
adsl <- random.cdisc.data::cadsl %>% slice(1:15)
adlb <- random.cdisc.data::cadlb %>% filter(USUBJID %in% adsl$USUBJID)
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
adlb <- df_explicit_na(adlb)
adlb_f <- adlb %>%
filter(
SAFFL == "Y",
PARAMCD == "ALT",
AVISIT != "SCREENING",
ARMCD == "ARM A"
) %>%
mutate(Patient_ID = sub(".*id-", "", USUBJID))
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
::::: panel-tabset
## Standard Plot
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
The user can select different `plotting_choices` depending on their preference. To demonstrate, separate plots are produced with a maximum of 3 observations each.
```{r plots1, test = list(plots_v1 = "plots")}
plots <- g_ipp(
df = adlb_f,
xvar = "AVISIT",
yvar = "AVAL",
xlab = "Visit",
ylab = "SGOT/ALT (U/L)",
id_var = "Patient_ID",
title = "Individual Patient Plots",
subtitle = "Treatment Arm A",
plotting_choices = "split_by_max_obs",
max_obs_per_plot = 3
)
plots
```
```{r include = FALSE}
webr_code_labels <- c("plots1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Plot with Patient Baselines as Reference
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
Here, patients' individual baseline values will be shown for reference. Note that users can provide their own custom theme to the function via the `ggtheme` argument.
```{r plots2, test = list(plots_v2 = "plots")}
plots <- g_ipp(
df = adlb_f,
xvar = "AVISIT",
yvar = "AVAL",
xlab = "Visit",
ylab = "SGOT/ALT (U/L)",
id_var = "Patient_ID",
title = "Individual Patient Plots",
subtitle = "Treatment Arm A",
add_baseline_hline = TRUE,
yvar_baseline = "BASE",
ggtheme = theme_minimal(),
plotting_choices = "split_by_max_obs",
max_obs_per_plot = 3
)
plots
```
```{r include = FALSE}
webr_code_labels <- c("plots2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
:::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.clinical)
## Data reproducible code
data <- teal_data()
data <- within(data, {
library(dplyr)
# use small sample size
ADSL <- random.cdisc.data::cadsl %>% slice(1:15)
ADLB <- random.cdisc.data::cadlb %>% filter(USUBJID %in% ADSL$USUBJID)
# Ensure character variables are converted to factors and empty strings and NAs are explicit missing levels.
ADSL <- df_explicit_na(ADSL)
ADLB <- df_explicit_na(ADLB) %>%
filter(AVISIT != "SCREENING")
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADLB")]
## Reusable Configuration For Modules
ADLB <- data[["ADLB"]]
## Setup App
app <- init(
data = data,
modules = modules(
tm_g_ipp(
label = "Individual Patient Plot",
dataname = "ADLB",
arm_var = choices_selected(
value_choices(ADLB, c("ARMCD")),
"ARM A"
),
paramcd = choices_selected(
value_choices(ADLB, "PARAMCD"),
"ALT"
),
aval_var = choices_selected(
variable_choices(ADLB, c("AVAL")),
"AVAL"
),
avalu_var = choices_selected(
variable_choices(ADLB, c("AVALU")),
"AVALU",
fixed = TRUE
),
id_var = choices_selected(
variable_choices(ADLB, c("USUBJID")),
"USUBJID",
fixed = TRUE
),
visit_var = choices_selected(
variable_choices(ADLB, c("AVISIT")),
"AVISIT"
),
baseline_var = choices_selected(
variable_choices(ADLB, c("BASE")),
"BASE",
fixed = TRUE
),
add_baseline_hline = FALSE,
separate_by_obs = FALSE
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}