---
title: BWG01
subtitle: Box Plot
---
------------------------------------------------------------------------
{{< include ../../_utils/envir_hook.qmd >}}
```{r setup, echo = FALSE, warning = FALSE, message = FALSE}
library(dplyr)
library(ggplot2)
library(nestcolor)
adlb <- random.cdisc.data::cadlb
adlb <- adlb %>% filter(PARAMCD == "ALT" & AVISIT == "WEEK 2 DAY 15")
# Definition of boxplot boundaries and whiskers
five_num <- function(x, probs = c(0, 0.25, 0.5, 0.75, 1)) {
r <- quantile(x, probs)
names(r) <- c("ymin", "lower", "middle", "upper", "ymax")
r
}
# get outliers based on quantile
# for outliers based on IQR see coef in geom_boxplot
get_outliers <- function(x, probs = c(0.05, 0.95)) {
r <- subset(x, x < quantile(x, probs[1]) | quantile(x, probs[2]) < x)
if (!is.null(x)) {
x_names <- subset(names(x), x < quantile(x, probs[1]) | quantile(x, probs[2]) < x)
names(r) <- x_names
}
r
}
# create theme used for all plots
theme_bp <- theme(
plot.title = element_text(hjust = 0),
plot.subtitle = element_text(hjust = 0),
plot.caption = element_text(hjust = 0),
panel.background = element_rect(fill = "white", color = "grey50")
)
# assign fill color and outline color
fc <- "#eaeef5"
oc <- getOption("ggplot2.discrete.fill")[1]
# get plot metadata data to derive coordinates for adding annotations
bp_annos <- function(bp, color, annos = 1) {
bp_mdat <- ggplot_build(bp)$data[[1]]
if (annos == 1) {
bp <- bp +
geom_segment(data = bp_mdat, aes(
x = xmin + (xmax - xmin) / 4, xend = xmax - (xmax - xmin) / 4,
y = ymax, yend = ymax
), linewidth = .5, color = color) +
geom_segment(data = bp_mdat, aes(
x = xmin + (xmax - xmin) / 4, xend = xmax - (xmax - xmin) / 4,
y = ymin, yend = ymin
), linewidth = .5, color = color)
} else {
bp <- bp +
geom_segment(data = bp_mdat, aes(
x = xmin + (xmax - xmin) / 4, xend = xmax - (xmax - xmin) / 4,
y = ymax, yend = ymax
), linewidth = .5, color = color) +
geom_segment(data = bp_mdat, aes(
x = xmin + (xmax - xmin) / 4, xend = xmax - (xmax - xmin) / 4,
y = ymin, yend = ymin
), linewidth = .5, color = color) +
geom_segment(data = bp_mdat, aes(
x = xmin, xend = xmax,
y = middle, yend = middle
), colour = color, linewidth = .5)
}
return(bp)
}
```
```{r include = FALSE}
webr_code_labels <- c("setup")
```
{{< include ../../_utils/webr_no_include.qmd >}}
## Output
:::::::::::: panel-tabset
## Standard Plot
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r plot1, test = list(plot_v1 = "plot")}
bp_1 <- ggplot(adlb, aes(x = ARMCD, y = AVAL)) +
stat_summary(geom = "boxplot", fun.data = five_num, fill = fc, color = oc) +
stat_summary(geom = "point", fun = mean, size = 3, shape = 8) +
labs(
title = "Box Plot of Laboratory Test Results",
subtitle = paste("Visit:", adlb$AVISIT[1]),
caption = "The whiskers extend to the minimum and maximum values.",
x = "Treatment Group",
y = paste0(adlb$PARAMCD[1], " (", adlb$AVALU[1], ")")
) +
theme_bp
plot <- bp_annos(bp_1, oc)
plot
```
```{r include = FALSE}
webr_code_labels <- c("plot1")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Plot Changing <br/> Whiskers
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r plot2, test = list(plot_v2 = "plot")}
bp_3 <- ggplot(adlb, aes(x = ARMCD, y = AVAL)) +
stat_summary(
geom = "boxplot", fun.data = five_num,
fun.args = list(probs = c(0.05, 0.25, 0.5, 0.75, 0.95)), fill = fc, color = oc
) +
stat_summary(geom = "point", fun = mean, size = 3, shape = 8) +
labs(
title = "Box Plot of Laboratory Test Results",
subtitle = paste("Visit:", adlb$AVISIT[1]),
caption = "The whiskers extend to the 5th and 95th percentile. Values outside the whiskers have not been plotted.",
x = "Treatment Group",
y = paste0(adlb$PARAMCD[1], " (", adlb$AVALU[1], ")")
) +
theme_bp
plot <- bp_annos(bp_3, oc)
plot
```
```{r include = FALSE}
webr_code_labels <- c("plot2")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Plot Adding <br/> Outliers
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r plot3, test = list(plot_v3 = "plot")}
bp_4 <- ggplot(adlb, aes(x = ARMCD, y = AVAL)) +
stat_summary(
geom = "boxplot", fun.data = five_num,
fun.args = list(probs = c(0.05, 0.25, 0.5, 0.75, 0.95)), fill = fc, color = oc
) +
stat_summary(geom = "point", fun = get_outliers, shape = 1) +
stat_summary(geom = "point", fun = mean, size = 3, shape = 8) +
labs(
title = "Box Plot of Laboratory Test Results",
subtitle = paste("Visit:", adlb$AVISIT[1]),
caption = "The whiskers extend to the 5th and 95th percentile.",
x = "Treatment Group",
y = paste0(adlb$PARAMCD[1], " (", adlb$AVALU[1], ")")
) +
theme_bp
plot <- bp_annos(bp_4, oc)
plot
```
```{r include = FALSE}
webr_code_labels <- c("plot3")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Plot Specifying Marker <br/> for Outliers and <br/> Adding Patient ID
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r plot4, test = list(plot_v4 = "plot")}
adlb_o <- adlb %>%
group_by(ARMCD) %>%
mutate(OUTLIER = AVAL < quantile(AVAL, 0.01) | AVAL > quantile(AVAL, 0.99)) %>%
filter(OUTLIER == TRUE) %>%
select(ARMCD, AVAL, USUBJID)
# Next step may be study-specific: shorten USUBJID to make annotation labels
# next 2 lines of code split USUBJID by "-" and take the last syllable as ID
n_split <- max(vapply(strsplit(adlb_o$USUBJID, "-"), length, numeric(1)))
adlb_o$ID <- vapply(strsplit(adlb_o$USUBJID, "-"), `[[`, n_split, FUN.VALUE = "a")
bp_5 <- ggplot(adlb, aes(x = ARMCD, y = AVAL)) +
stat_summary(
geom = "boxplot", fun.data = five_num,
fun.args = list(probs = c(0.01, 0.25, 0.5, 0.75, 0.99)), fill = fc, color = oc
) +
stat_summary(geom = "point", fun = mean, size = 3, shape = 8) +
geom_point(data = adlb_o, aes(x = ARMCD, y = AVAL), shape = 1) +
geom_text(data = adlb_o, aes(x = ARMCD, y = AVAL, label = ID), size = 3, hjust = -0.2) +
labs(
title = "Box Plot of Laboratory Test Results",
subtitle = paste("Visit:", adlb$AVISIT[1]),
caption = "The whiskers extend to the largest and smallest observed value within 1.5*IQR.",
x = "Treatment Group",
y = paste0(adlb$PARAMCD[1], " (", adlb$AVALU[1], ")")
) +
theme_bp
plot <- bp_annos(bp_5, oc)
plot
```
```{r include = FALSE}
webr_code_labels <- c("plot4")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Plot Specifying <br/> Marker for Mean
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r plot5, test = list(plot_v5 = "plot")}
bp_6 <- ggplot(adlb, aes(x = ARMCD, y = AVAL)) +
geom_boxplot(fill = fc, color = oc) +
stat_summary(geom = "point", fun = mean, size = 3, shape = 5) +
labs(
title = "Box Plot of Laboratory Test Results",
subtitle = paste("Visit:", adlb$AVISIT[1]),
caption = "The whiskers extend to the minimum and maximum values.",
x = "Treatment Group",
y = paste0(adlb$PARAMCD[1], " (", adlb$AVALU[1], ")")
) +
theme_bp
plot <- bp_annos(bp_6, oc)
plot
```
```{r include = FALSE}
webr_code_labels <- c("plot5")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Plot by Treatment <br/> and Timepoint
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r plot6, test = list(plot_v6 = "plot")}
adsl <- random.cdisc.data::cadsl
adlb <- random.cdisc.data::cadlb
adlb_v <- adlb %>%
filter(PARAMCD == "ALT" & AVISIT %in% c("WEEK 1 DAY 8", "WEEK 2 DAY 15", "WEEK 3 DAY 22", "WEEK 4 DAY 29"))
bp_7 <- ggplot(adlb_v, aes(x = AVISIT, y = AVAL)) +
stat_summary(
geom = "boxplot",
fun.data = five_num,
position = position_dodge2(.5),
aes(fill = ARMCD, color = ARMCD)
) +
stat_summary(
geom = "point",
fun = mean,
aes(group = ARMCD),
size = 3,
shape = 8,
position = position_dodge2(1)
) +
labs(
title = "Box Plot of Laboratory Test Results",
caption = "The whiskers extend to the minimum and maximum values.",
x = "Visit",
y = paste0(adlb$PARAMCD[1], " (", adlb$AVALU[1], ")")
) +
theme_bp
plot <- bp_annos(bp_7, oc, 2)
plot
```
```{r include = FALSE}
webr_code_labels <- c("plot6")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Plot by Timepoint <br/> and Treatment
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r plot7, test = list(plot_v7 = "plot")}
bp_8 <- ggplot(adlb_v, aes(x = ARMCD, y = AVAL)) +
stat_summary(
geom = "boxplot",
fun.data = five_num,
position = position_dodge2(width = .5),
aes(fill = AVISIT, color = AVISIT)
) +
stat_summary(
geom = "point",
fun = mean,
aes(group = AVISIT),
size = 3,
shape = 8,
position = position_dodge2(1)
) +
labs(
title = "Box Plot of Laboratory Test Results",
caption = "The whiskers extend to the minimum and maximum values.",
x = "Treatment Group",
y = paste0(adlb$PARAMCD[1], " (", adlb$AVALU[1], ")")
) +
theme_bp
plot <- bp_annos(bp_8, oc, 2)
plot
```
```{r include = FALSE}
webr_code_labels <- c("plot7")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Plot with <br/> Table Section
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r plot8, test = list(plot_v8 = "plot")}
# Make wide dataset with summary statistics
adlb_wide <- adlb %>%
group_by(ARMCD) %>%
summarise(
n = n(),
mean = round(mean(AVAL), 1),
median = round(median(AVAL), 1),
median_ci = paste(round(DescTools::MedianCI(AVAL)[1:2], 1), collapse = " - "),
q1_q3 = paste(round(quantile(AVAL, c(0.25, 0.75)), 1), collapse = " - "),
min_max = paste0(round(min(AVAL), 1), "-", max(round(AVAL, 2)))
)
# Make long dataset
adlb_long <- tidyr::gather(adlb_wide, key = type, value = stat, n:min_max)
adlb_long <- adlb_long %>% mutate(
type_lbl = case_when(
type == "n" ~ "n",
type == "mean" ~ "Mean",
type == "median" ~ "Median",
type == "median_ci" ~ "95% CI for Median",
type == "q1_q3" ~ "25% and 75%-ile",
type == "min_max" ~ "Min - Max"
)
)
adlb_long$type_lbl <- factor(adlb_long$type_lbl,
levels = c("Min - Max", "25% and 75%-ile", "95% CI for Median", "Median", "Mean", "n")
)
bp_9 <- ggplot(adlb, aes(x = ARMCD, y = AVAL)) +
stat_summary(geom = "boxplot", fun.data = five_num, fill = fc, color = oc) +
stat_summary(geom = "point", fun = mean, size = 3, shape = 8) +
labs(
title = "Box Plot of Laboratory Test Results",
subtitle = paste("Visit:", adlb$AVISIT[1]),
x = "Treatment Group",
y = paste0(adlb$PARAMCD[1], " (", adlb$AVALU[1], ")")
) +
theme_bp
tbl_theme <- theme(
panel.border = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.ticks = element_blank(),
axis.title = element_blank(),
axis.text.y = element_text(face = "plain"),
axis.text.x = element_blank()
)
tbl_1 <- ggplot(adlb_long, aes(x = ARMCD, y = type_lbl, label = stat), vjust = 10) +
geom_text(size = 3) +
scale_y_discrete(labels = levels(adlb_long$type_lbl)) +
theme_bw() +
tbl_theme +
labs(caption = "The whiskers extend to the minimum and maximum values.") +
theme_bp
plot <- cowplot::plot_grid(bp_annos(bp_9, oc), tbl_1,
rel_heights = c(4, 2),
ncol = 1, nrow = 2, align = "v"
)
plot
```
```{r include = FALSE}
webr_code_labels <- c("plot8")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Plot with Number of Patients <br/> only in Table Section
::: {.panel-tabset .nav-justified group="webr"}
## {{< fa regular file-lines sm fw >}} Preview
```{r plot9, test = list(plot_v9 = "plot")}
tbl_2 <- adlb_long %>%
filter(type == "n") %>%
ggplot(aes(x = ARMCD, y = type_lbl, label = stat)) +
geom_text(size = 3) +
scale_y_discrete(labels = "n") +
theme_bw() +
tbl_theme +
labs(caption = "The whiskers extend to the minimum and maximum values.") +
theme_bp
plot <- cowplot::plot_grid(bp_annos(bp_9, oc), tbl_2,
rel_heights = c(6, 1),
ncol = 1, nrow = 2, align = "v"
)
plot
```
```{r include = FALSE}
webr_code_labels <- c("plot9")
```
{{< include ../../_utils/webr.qmd >}}
:::
## Data Setup
```{r setup}
#| code-fold: show
```
::::::::::::
{{< include ../../_utils/save_results.qmd >}}
## `teal` App
::: {.panel-tabset .nav-justified}
## {{< fa regular file-lines fa-sm fa-fw >}} Preview
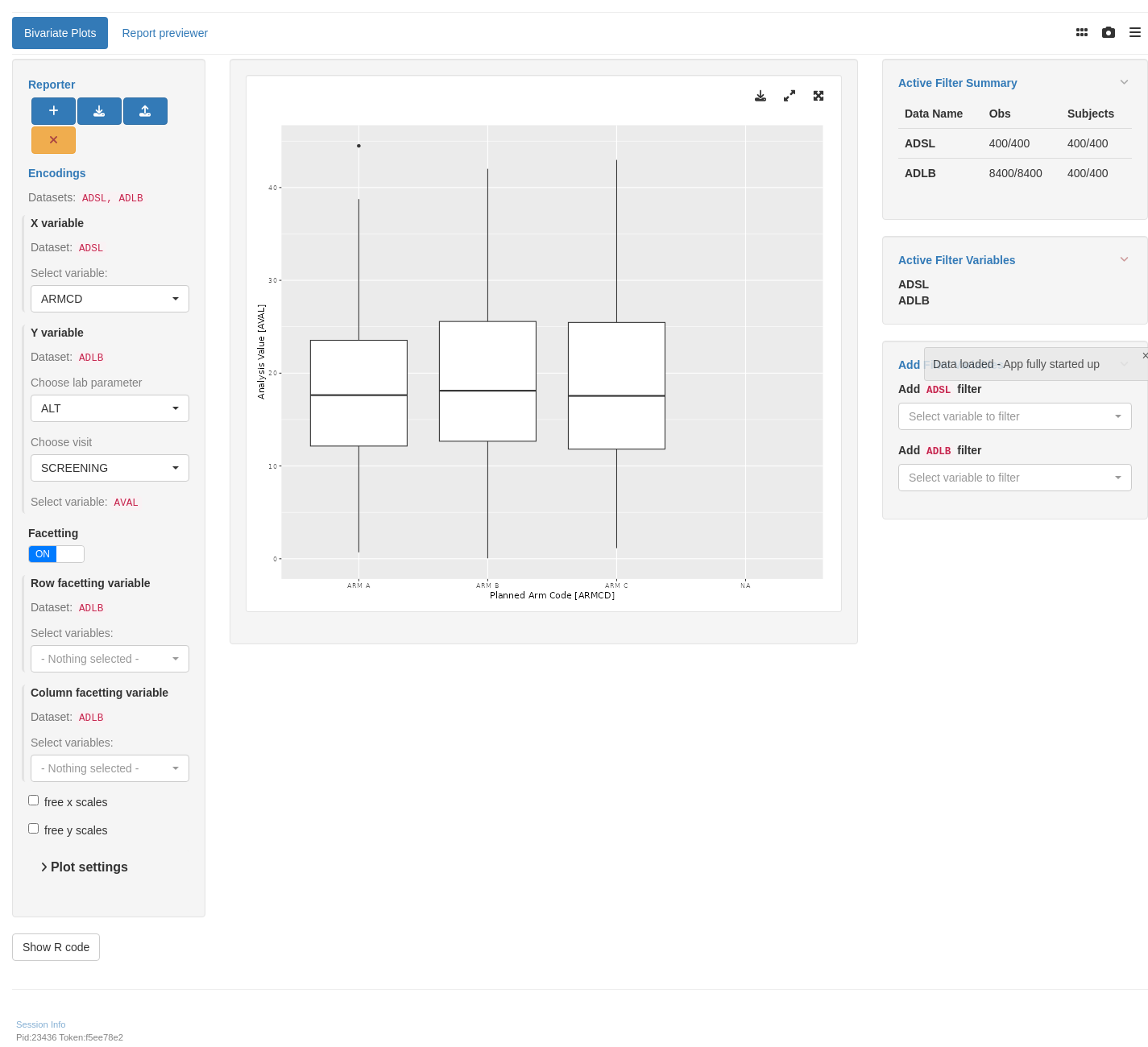
```{r teal, opts.label = c("skip_if_testing", "app")}
library(teal.modules.general)
## Data reproducible code
data <- teal_data()
data <- within(data, {
library(tern)
ADSL <- random.cdisc.data::cadsl
ADLB <- random.cdisc.data::cadlb
# If PARAMCD and AVISIT are not factors, convert to factors
# Also fill in missing values with "<Missing>"
ADLB <- ADLB %>%
df_explicit_na(
omit_columns = setdiff(names(ADLB), c("PARAMCD", "AVISIT")),
char_as_factor = TRUE
)
# If statment below fails, pre-process ADLB to be one record per
# study, subject, param and visit eg. filter with ANLFL = 'Y'
stopifnot(nrow(ADLB) == nrow(unique(ADLB[, c("STUDYID", "USUBJID", "PARAMCD", "AVISIT")])))
})
join_keys(data) <- default_cdisc_join_keys[c("ADSL", "ADLB")]
## Reusable Configuration For Modules
ADSL <- data[["ADSL"]]
ADLB <- data[["ADLB"]]
## Setup App
app <- init(
data = data,
modules = modules(
tm_g_bivariate(
x = data_extract_spec(
dataname = "ADSL",
select = select_spec(
label = "Select variable:",
choices = names(ADSL),
selected = "ARMCD",
fixed = FALSE
)
),
y = data_extract_spec(
dataname = "ADLB",
filter = list(
filter_spec(
vars = c("PARAMCD"),
choices = levels(ADLB$PARAMCD),
selected = levels(ADLB$PARAMCD)[1],
multiple = FALSE,
label = "Choose lab parameter"
),
filter_spec(
vars = c("AVISIT"),
choices = levels(ADLB$AVISIT),
selected = levels(ADLB$AVISIT)[1],
multiple = FALSE,
label = "Choose visit"
)
),
select = select_spec(
label = "Select variable:",
choices = names(ADLB),
selected = "AVAL",
multiple = FALSE,
fixed = TRUE
)
),
row_facet = data_extract_spec(
dataname = "ADLB",
select = select_spec(
label = "Select variables:",
choices = names(ADLB),
selected = NULL,
multiple = FALSE,
fixed = FALSE
)
),
col_facet = data_extract_spec(
dataname = "ADLB",
select = select_spec(
label = "Select variables:",
choices = names(ADLB),
selected = NULL,
multiple = FALSE,
fixed = FALSE
)
)
)
)
)
shinyApp(app$ui, app$server)
```
{{< include ../../_utils/shinylive.qmd >}}
:::
{{< include ../../repro.qmd >}}