SFG3
Survival Forest Graphs for One Treatment Arm
We prepare the data similarly as in SFG1, focusing on a single arm in the biomarker evaluable population.
We define a vector of all cutpoints to use for a numeric biomarker (here BMRKR1). We lapply() over this vector, each time generating a binary factor variable BMRKR1_cut and then tabulating the resulting statistics as e.g. in SFG1. Then we rbind() all tables in the list together.
Code
all_cutpoints <- c(2.5, 5, 7.5, 10)
tables_all_cutpoints <- lapply(all_cutpoints, function(cutpoint) {
adtte_cut <- adtte %>%
mutate(
BMRKR1_cut = explicit_na(factor(
ifelse(BMRKR1 > cutpoint, "Greater", "Less")
))
)
tbl <- extract_survival_subgroups(
variables = list(
tte = "AVAL",
is_event = "is_event",
arm = "BMRKR1_cut"
),
label_all = paste0("BMRKR1 (", cutpoint, ")"),
data = adtte_cut
)
basic_table() %>%
tabulate_survival_subgroups(
df = tbl,
vars = c("n_tot_events", "n", "n_events", "median", "hr", "ci"),
time_unit = adtte_cut$AVALU[1]
)
})
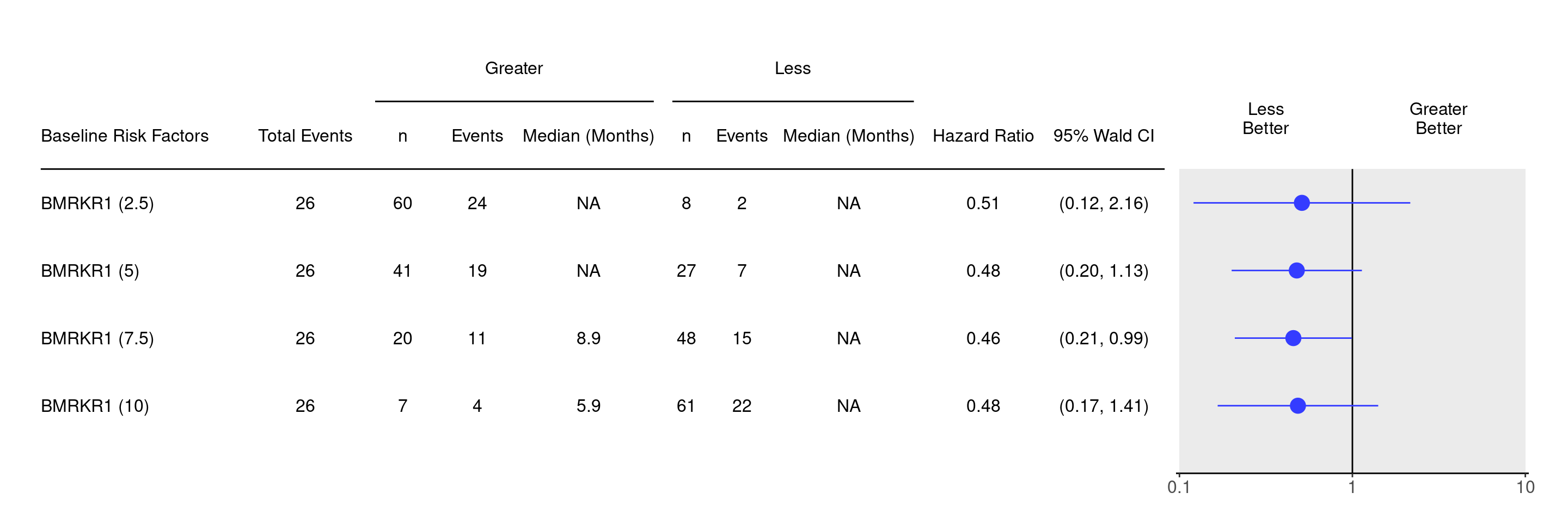
result <- do.call(rbind, tables_all_cutpoints)We can now produce the forest plot using the g_forest() function. Notice that the result object in these derivation steps has lost its attributes. In order to specify the column indices for the estimator, confidence interval, and header, we need to derive the attributes from one of the original tables and specify them using the col_x, col_y and forest_header arguments of g_forest().
R version 4.4.1 (2024-06-14)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_1.1.4 tern_0.9.5.9022 rtables_0.6.9.9014
[4] magrittr_2.0.3 formatters_0.5.9.9001
loaded via a namespace (and not attached):
[1] Matrix_1.7-0 gtable_0.3.5
[3] jsonlite_1.8.8 compiler_4.4.1
[5] tidyselect_1.2.1 stringr_1.5.1
[7] tidyr_1.3.1 splines_4.4.1
[9] scales_1.3.0 yaml_2.3.10
[11] fastmap_1.2.0 lattice_0.22-6
[13] ggplot2_3.5.1 R6_2.5.1
[15] labeling_0.4.3 generics_0.1.3
[17] knitr_1.48 forcats_1.0.0
[19] rbibutils_2.2.16 htmlwidgets_1.6.4
[21] backports_1.5.0 checkmate_2.3.2
[23] tibble_3.2.1 munsell_0.5.1
[25] pillar_1.9.0 rlang_1.1.4
[27] utf8_1.2.4 broom_1.0.6
[29] stringi_1.8.4 xfun_0.47
[31] cli_3.6.3 withr_3.0.1
[33] Rdpack_2.6.1 digest_0.6.37
[35] grid_4.4.1 cowplot_1.1.3
[37] lifecycle_1.0.4 vctrs_0.6.5
[39] evaluate_0.24.0 glue_1.7.0
[41] farver_2.1.2 codetools_0.2-20
[43] survival_3.7-0 random.cdisc.data_0.3.15.9009
[45] fansi_1.0.6 colorspace_2.1-1
[47] purrr_1.0.2 rmarkdown_2.28
[49] tools_4.4.1 pkgconfig_2.0.3
[51] htmltools_0.5.8.1