DG1B
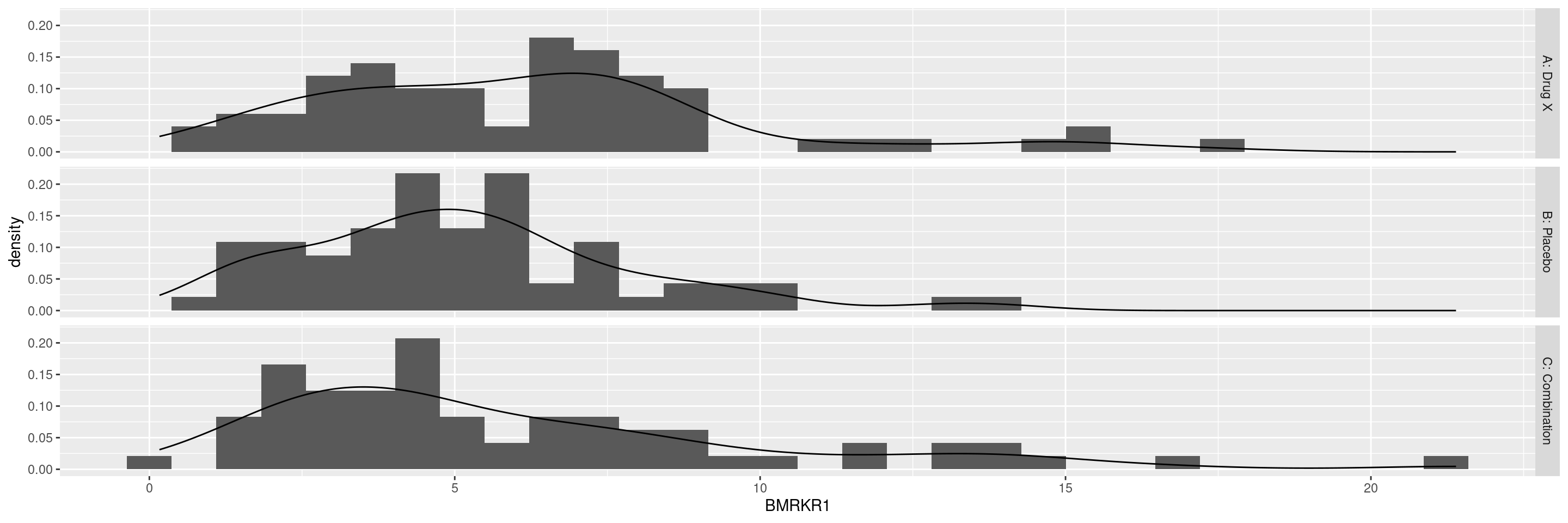
Histogram of One Numeric Variable by Treatment Arm
DG
We will use the cadsl data set from the random.cdisc.data package and ggplot2 to create the plots. In this example, we will plot histograms of one or multiple numeric variables. We start by selecting the biomarker evaluable population with the flag variable BEP01FL and then populating a new continuous biomarker variable, BMRKR3.
Below example shows histograms for the BMRKR1 variable by treatment ARM. Including a statistics table in this graph works in the same way as we did above for DG1A.
R version 4.4.1 (2024-06-14)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] tidyr_1.3.1 tibble_3.2.1 dplyr_1.1.4
[4] ggplot2.utils_0.3.2 ggplot2_3.5.1 tern_0.9.5.9022
[7] rtables_0.6.9.9014 magrittr_2.0.3 formatters_0.5.9.9001
loaded via a namespace (and not attached):
[1] utf8_1.2.4 generics_0.1.3
[3] EnvStats_3.0.0 stringi_1.8.4
[5] lattice_0.22-6 digest_0.6.37
[7] evaluate_0.24.0 grid_4.4.1
[9] fastmap_1.2.0 jsonlite_1.8.8
[11] Matrix_1.7-0 backports_1.5.0
[13] survival_3.7-0 purrr_1.0.2
[15] fansi_1.0.6 scales_1.3.0
[17] codetools_0.2-20 Rdpack_2.6.1
[19] cli_3.6.3 ggpp_0.5.8-1
[21] rlang_1.1.4 rbibutils_2.2.16
[23] munsell_0.5.1 splines_4.4.1
[25] withr_3.0.1 yaml_2.3.10
[27] tools_4.4.1 polynom_1.4-1
[29] checkmate_2.3.2 colorspace_2.1-1
[31] forcats_1.0.0 ggstats_0.6.0
[33] broom_1.0.6 vctrs_0.6.5
[35] R6_2.5.1 lifecycle_1.0.4
[37] stringr_1.5.1 htmlwidgets_1.6.4
[39] MASS_7.3-61 pkgconfig_2.0.3
[41] pillar_1.9.0 gtable_0.3.5
[43] glue_1.7.0 xfun_0.47
[45] tidyselect_1.2.1 knitr_1.48
[47] farver_2.1.2 htmltools_0.5.8.1
[49] labeling_0.4.3 rmarkdown_2.28
[51] random.cdisc.data_0.3.15.9009 compiler_4.4.1 Reuse
Copyright 2023, Hoffmann-La Roche Ltd.