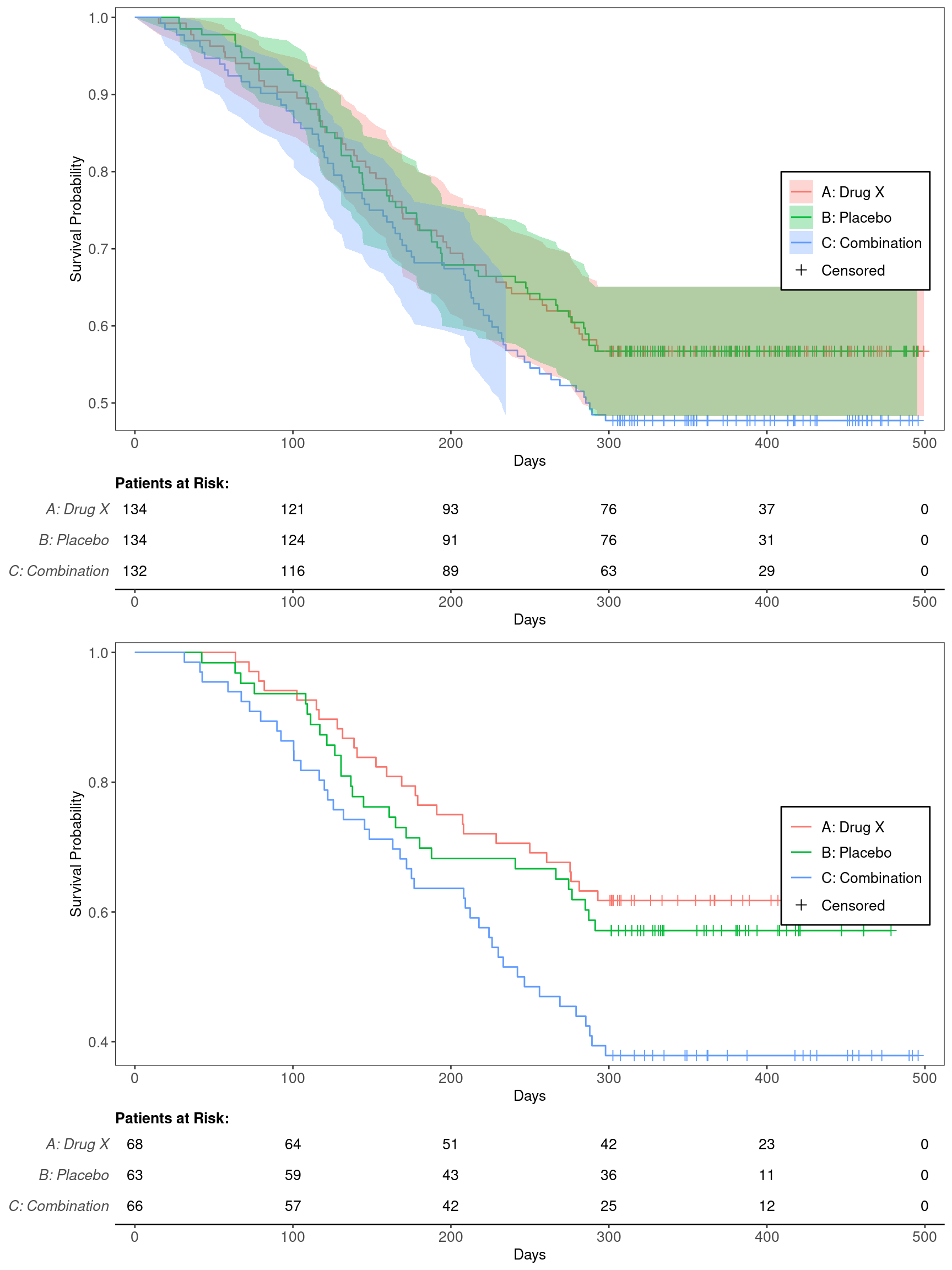
KG1B
Kaplan-Meier Graph for Comparing ITT and BEP Populations
We will use the cadtte data set from the random.cdisc.data package to create the Kaplan-Meier (KM) plots. We start by filtering the time-to-event dataset for the overall survival observations and by one treatment arm (A), creating a new variable for event information, and curating a list of variables required to produce the plot.
Here we only filter the time-to-event dataset for the overall survival observations, but keep all treatment arms and the overall population.
Plot the ITT (top, with CIs) and BEP (bottom, without CIs) KM graphs, convert them to ggplotGrob objects, and combine the plots via rbind. Then, use the grid package to create an empty plot area, and draw the KM curves on the graph device.
Code
R version 4.4.1 (2024-06-14)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] ggplot2_3.5.1 dplyr_1.1.4 tern_0.9.5.9022
[4] rtables_0.6.9.9014 magrittr_2.0.3 formatters_0.5.9.9001
loaded via a namespace (and not attached):
[1] Matrix_1.7-0 gtable_0.3.5
[3] jsonlite_1.8.8 compiler_4.4.1
[5] tidyselect_1.2.1 stringr_1.5.1
[7] tidyr_1.3.1 splines_4.4.1
[9] scales_1.3.0 yaml_2.3.10
[11] fastmap_1.2.0 lattice_0.22-6
[13] R6_2.5.1 labeling_0.4.3
[15] generics_0.1.3 knitr_1.48
[17] forcats_1.0.0 rbibutils_2.2.16
[19] htmlwidgets_1.6.4 backports_1.5.0
[21] checkmate_2.3.2 tibble_3.2.1
[23] munsell_0.5.1 pillar_1.9.0
[25] rlang_1.1.4 utf8_1.2.4
[27] broom_1.0.6 stringi_1.8.4
[29] xfun_0.47 cli_3.6.3
[31] withr_3.0.1 Rdpack_2.6.1
[33] digest_0.6.37 cowplot_1.1.3
[35] lifecycle_1.0.4 vctrs_0.6.5
[37] evaluate_0.24.0 glue_1.7.0
[39] farver_2.1.2 codetools_0.2-20
[41] survival_3.7-0 random.cdisc.data_0.3.15.9009
[43] fansi_1.0.6 colorspace_2.1-1
[45] purrr_1.0.2 rmarkdown_2.28
[47] tools_4.4.1 pkgconfig_2.0.3
[49] htmltools_0.5.8.1