Introduction
The purpose of this vignette is to explain the use of
getClinicalTrials(), the core function of this package.
getClinicalTrials() can be used to simulate a large number
of clinical trials with endpoints progression-free survival (PFS) and
overall survival (OS). The underlying model is a survival multistate
model with an initial state, an intermediate progression
state and an absorbing death state. The multistate model
approach has the advantage of naturally accounting for the correlation
between the PFS and OS endpoints (Meller,
Beyersmann, and Rufibach 2019). OS is defined as the time to
reach the absorbing state death and PFS is defined as the time to reach
the absorbing state death or the intermediate state
progression, whichever occurs first. Figure 1 shows the
multistate model with the corresponding transition hazards. To get
started quickly, we show a simple and quick example of a clinical trial
simulation.
#> Warning in plot.Hist(idHist, stateLabels = stateLabels, box1.row = 2, box1.column = 1, : The dimension of the boxes may depend on the current graphical device
#> in the sense that the layout and centering of text may change when you resize the graphical device and call the same plot.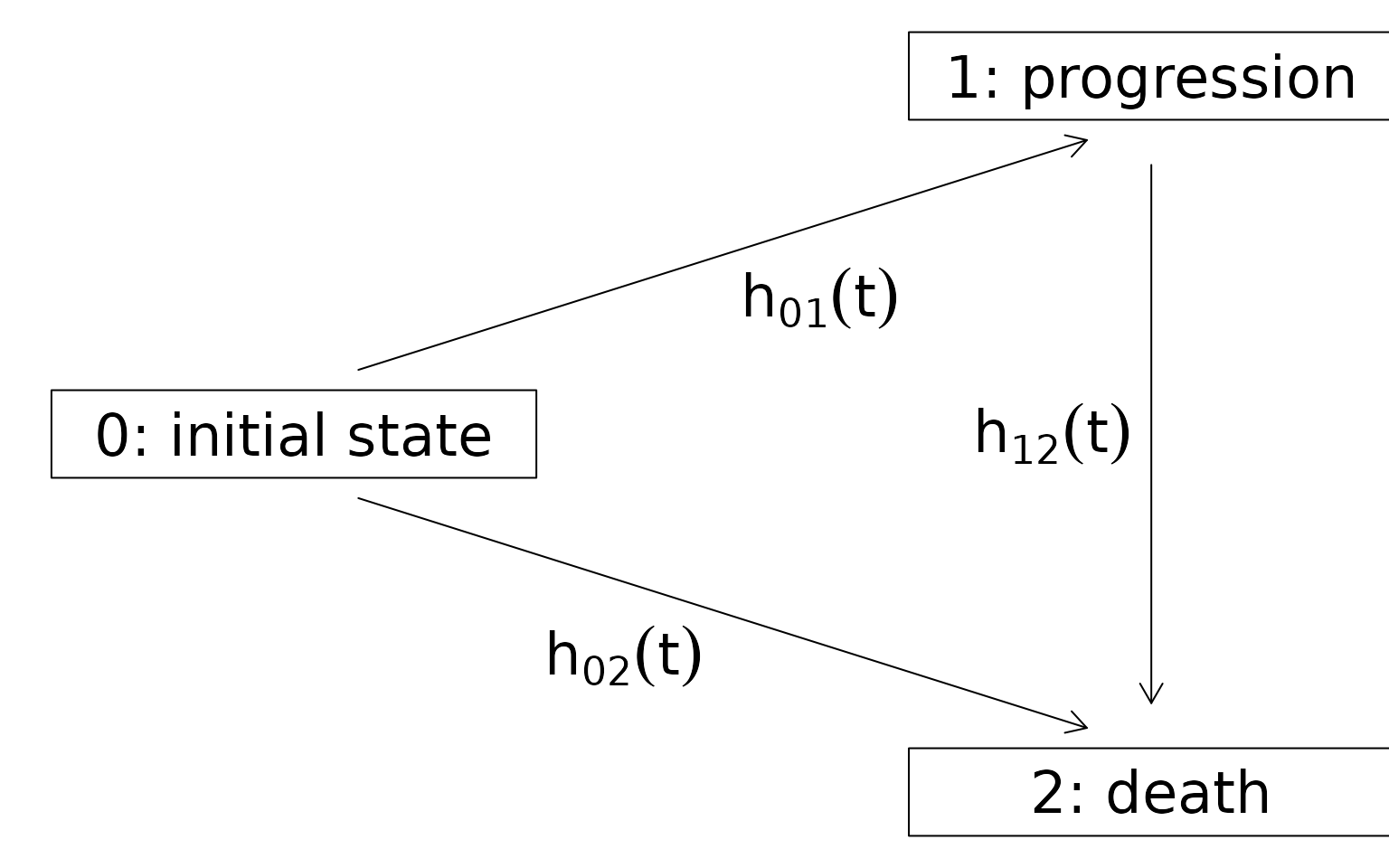
Figure 1 - Multistate model with indermediate state progession and absorbing state death
Simulation Specifications
getClinicalTrials() simulates trials with an arbitrary
number of treatment arms. In general, the argument specifications were
tried to be similar to the R-package Rpact (Wassmer and Pahlke 2021) in order to be more
user-friendly. For illustration, we consider a trial with two arms. The
number of patients per trial must be specified. In our example
simulation there are 30 patients in the first treatment arm and 60 in
the second treatment arm:
nPat <- c(30, 60)The transition parameters describing the transition hazards per
treatment arm must be passed as a list to
getClinicalTrials(). The class
TransitionParameters contains the elements
hazards, intervals, weibull_rates
and family which allow to describe transition hazards
leading to exponential, piecewise exponential or Weibull distributed
survival times. There are some helper functions in the
simIDM package to create the appropriate transition
parameters for the desired distribution. -
exponential_transition(): creates a list with class
TransitionParameters containing hazards, time intervals and
Weibull rates for exponential event times in an illness-death model. -
piecewise_exponential(): creates a list with class
TransitionParameters containing hazards, time intervals and
Weibull rates for piecewise exponential event times in an illness-death
model. - weibull_transition(): creates a list with class
TransitionParameters containing hazards, time intervals and
Weibull rates for Weibull distributed event times in an illness-death
model.
In our example we consider constant hazard rates, i.e. exponential distributed event times.
library(simIDM)
transitionGroup1 <- exponential_transition(h01 = 1.2, h02 = 1.5, h12 = 1.6)
print(transitionGroup1$hazards)
#> $h01
#> [1] 1.2
#>
#> $h02
#> [1] 1.5
#>
#> $h12
#> [1] 1.6
transitionGroup2 <- exponential_transition(h01 = 1, h02 = 1.3, h12 = 1.7)
print(transitionGroup2$hazards)
#> $h01
#> [1] 1
#>
#> $h02
#> [1] 1.3
#>
#> $h12
#> [1] 1.7
transitionbyArm <- list(transitionGroup1, transitionGroup2)The specification for random patient dropouts can be passed directly
to getClinicalTrials(). A list with elements
rate and time specifies the drop-out
probability. Random censoring times are generated using the exponential
distribution. dropout$rate specifies the drop-out
probability per dropout$time time units. The dropout
parameters can be specified either as one list that should be applied to
all treatment groups or a separate list for each treatment group. If
dropout$rate is equal to 0, then no censoring due to
dropout is applied. In our example we expect that 10% of the patients
drop-out within 12 months per treatment group (we take month as the time
unit of our trial), thus we specify:
dropout <- list(rate = 0.1, time = 12)There are two time-scales that we need to consider when simulating
multistate data. The individual time scale with start in the initial
state at time 0 and the study time scale, which matches the individual
time scale only if the study start time (e.g. randomization date) is the
same time in calendar time for all patients. It can also be specified
that patients enter the study in a staggered manner. Uniformly
distributed random variables are used to generate staggered study entry
times. There are two possibilities for the specification: Either, the
length of the accrual period is specified, e.g. 12 month, in which case
the staggered entry times are generated in each treatment group using
random variables \(U ~Unif(0, 12)\).
Or, it is specified how many patients are recruited per time unit in
each treatment group. Thus, if 10 patients are recruited per month, then
the staggered entry times are generated using random variables \(U ~Unif(0, 30/10)\). A list
accrual with elements accrualParam and
accrualValue is passed to getClinicalTrials()
describing the accrual intensity. For accrualParam equal
time, accrualValuedescribes the length of the accrual
period. For accrualParam equal intensity, it describes the
number of patients recruited per time unit. If accrualValue
is equal to 0, all patients start at the same calender time in the
initial state. The accrual parameters can be specified either as one
list that should be applied to all treatment groups or a separate list
for each treatment group.
Application
We now simulate 100 (nRep=100) clinical trials with the
parameters described and specified above. There are two options for how
the output data is presented, one row per patient or one row per
transition and patient. Let’s take a look at both.
simStudies1 <- getClinicalTrials(
nRep = 100, nPat = nPat, seed = 1234, datType = "1rowTransition",
transitionByArm = transitionbyArm, dropout = dropout,
accrual = accrual
)
#> Simulating 100 trials:
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
simStudies2 <- getClinicalTrials(
nRep = 100, nPat = nPat, seed = 1234, datType = "1rowPatient",
transitionByArm = transitionbyArm, dropout = dropout,
accrual = accrual
)
#> Simulating 100 trials:
#>
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%Output Data
We get as output a list with nRep elements, each
containing a data set of a single simulated trial. The specification of
the seed ensures, that the the datasets of the two lists contain the
same study data, only the presentation is different.
simStudies1 shows the transition times per patient and
simStudies2 the PFS and OS times per patient.
head(simStudies1[[1]], 6)
#> id from to entry exit entryAct exitAct censAct trt
#> 1 1 0 2 0.0000000 0.46787743 1.234711 1.702588 286.171654 1
#> 2 2 0 2 0.0000000 0.26010392 7.971605 8.231709 36.076123 1
#> 3 3 0 2 0.0000000 0.06150124 7.442772 7.504273 8.192422 1
#> 4 4 0 1 0.0000000 0.25964639 9.159742 9.419389 207.649214 1
#> 5 4 1 2 0.2596464 2.13580247 9.419389 11.295545 207.649214 1
#> 6 5 0 2 0.0000000 0.25227459 9.945982 10.198257 54.044011 1
head(simStudies2[[1]], 5)
#> id trt PFStime CensoredPFS PFSevent OStime CensoredOS OSevent
#> 1 1 1 0.46787743 0 1 0.46787743 0 1
#> 2 2 1 0.26010392 0 1 0.26010392 0 1
#> 3 3 1 0.06150124 0 1 0.06150124 0 1
#> 4 4 1 0.25964639 0 1 2.13580247 0 1
#> 5 5 1 0.25227459 0 1 0.25227459 0 1
#> recruitTime OStimeCal PFStimeCal
#> 1 1.234711 1.702588 1.702588
#> 2 7.971605 8.231709 8.231709
#> 3 7.442772 7.504273 7.504273
#> 4 9.159742 11.295545 9.419389
#> 5 9.945982 10.198257 10.198257If we want to simulate only one trial,
getOneClinicalTrial can also be used and with
getDatasetWideFormat can be used to convert the dataset
from being displayed with transition time to displaying PFS and OS
times.
simStudy_Trans <- getOneClinicalTrial(
nPat = nPat, transitionByArm = transitionbyArm,
dropout = dropout,
accrual = accrual
)
simStudy_PFSOS <- getDatasetWideFormat(simStudy_Trans)
head(simStudy_Trans, 10)
#> id from to entry exit entryAct exitAct censAct trt
#> 1 1 0 2 0.0000000 0.32181077 5.063864 5.385675 335.59100 1
#> 2 2 0 1 0.0000000 0.44263317 8.083607 8.526241 96.83774 1
#> 3 2 1 2 0.4426332 0.70429214 8.526241 8.787899 96.83774 1
#> 4 3 0 2 0.0000000 0.03497509 4.478954 4.513929 74.19954 1
#> 5 4 0 1 0.0000000 0.19827637 5.606922 5.805199 48.06033 1
#> 6 4 1 2 0.1982764 0.93065829 5.805199 6.537580 48.06033 1
#> 7 5 0 2 0.0000000 0.06933912 9.077324 9.146663 119.07514 1
#> 8 6 0 1 0.0000000 0.44623034 8.790901 9.237131 81.78121 1
#> 9 6 1 2 0.4462303 0.44837021 9.237131 9.239271 81.78121 1
#> 10 7 0 1 0.0000000 0.63135991 8.441810 9.073170 97.36028 1
head(simStudy_PFSOS, 10)
#> id trt PFStime CensoredPFS PFSevent OStime CensoredOS OSevent
#> 1 1 1 0.32181077 0 1 0.32181077 0 1
#> 2 2 1 0.44263317 0 1 0.70429214 0 1
#> 3 3 1 0.03497509 0 1 0.03497509 0 1
#> 4 4 1 0.19827637 0 1 0.93065829 0 1
#> 5 5 1 0.06933912 0 1 0.06933912 0 1
#> 6 6 1 0.44623034 0 1 0.44837021 0 1
#> 7 7 1 0.63135991 0 1 1.69430412 0 1
#> 8 8 1 1.60671207 0 1 1.60671207 0 1
#> 9 9 1 0.22975563 0 1 0.50776145 0 1
#> 10 10 1 0.06312064 0 1 0.06312064 0 1
#> recruitTime OStimeCal PFStimeCal
#> 1 5.0638640 5.385675 5.385675
#> 2 8.0836073 8.787899 8.526241
#> 3 4.4789541 4.513929 4.513929
#> 4 5.6069222 6.537580 5.805199
#> 5 9.0773241 9.146663 9.146663
#> 6 8.7909011 9.239271 9.237131
#> 7 8.4418100 10.136114 9.073170
#> 8 0.4754946 2.082207 2.082207
#> 9 2.8876069 3.395368 3.117363
#> 10 6.6150703 6.678191 6.678191It is also possible to generate a a study that censors all patients after a pre-specified number of events (PFS or OS) occurred.
simStudy_Trans <- getOneClinicalTrial(
nPat = nPat, transitionByArm = transitionbyArm,
dropout = dropout,
accrual = accrual
)
simStudy_PFSOS <- getDatasetWideFormat(simStudy_Trans)
head(simStudy_Trans, 10)
#> id from to entry exit entryAct exitAct censAct trt
#> 1 1 0 1 0.00000000 0.06961555 7.780312062 7.8499276 102.95744 1
#> 2 1 1 2 0.06961555 1.04718403 7.849927611 8.8274961 102.95744 1
#> 3 2 0 2 0.00000000 0.26116360 4.626272118 4.8874357 47.17874 1
#> 4 3 0 2 0.00000000 0.15075060 6.508751956 6.6595026 168.99387 1
#> 5 4 0 1 0.00000000 0.56212147 2.690053477 3.2521749 48.75687 1
#> 6 4 1 2 0.56212147 2.41915162 3.252174950 5.1092051 48.75687 1
#> 7 5 0 2 0.00000000 0.01950124 8.332294915 8.3517962 118.78556 1
#> 8 6 0 2 0.00000000 0.47629626 0.024984612 0.5012809 145.10228 1
#> 9 7 0 2 0.00000000 0.64391257 0.003990494 0.6479031 63.08757 1
#> 10 8 0 2 0.00000000 0.56831986 7.120901188 7.6892211 659.17510 1
head(simStudy_PFSOS, 10)
#> id trt PFStime CensoredPFS PFSevent OStime CensoredOS OSevent
#> 1 1 1 0.06961555 0 1 1.04718403 0 1
#> 2 2 1 0.26116360 0 1 0.26116360 0 1
#> 3 3 1 0.15075060 0 1 0.15075060 0 1
#> 4 4 1 0.56212147 0 1 2.41915162 0 1
#> 5 5 1 0.01950124 0 1 0.01950124 0 1
#> 6 6 1 0.47629626 0 1 0.47629626 0 1
#> 7 7 1 0.64391257 0 1 0.64391257 0 1
#> 8 8 1 0.56831986 0 1 0.56831986 0 1
#> 9 9 1 0.27386179 0 1 0.27386179 0 1
#> 10 10 1 0.41250249 0 1 1.15933490 0 1
#> recruitTime OStimeCal PFStimeCal
#> 1 7.780312062 8.8274961 7.8499276
#> 2 4.626272118 4.8874357 4.8874357
#> 3 6.508751956 6.6595026 6.6595026
#> 4 2.690053477 5.1092051 3.2521749
#> 5 8.332294915 8.3517962 8.3517962
#> 6 0.024984612 0.5012809 0.5012809
#> 7 0.003990494 0.6479031 0.6479031
#> 8 7.120901188 7.6892211 7.6892211
#> 9 5.236761118 5.5106229 5.5106229
#> 10 5.930290024 7.0896249 6.3427925
simStudy_40PFS <- censoringByNumberEvents(data = simStudy_PFSOS, eventNum = 40, typeEvent = "PFS")
simStudy_30POS <- censoringByNumberEvents(data = simStudy_PFSOS, eventNum = 30, typeEvent = "PFS")
simStudy_40PFS
#> id trt PFStime PFSevent OStime CensoredOS OSevent recruitTime
#> 1 2 1 0.26116360 1 0.26116360 0 1 4.626272118
#> 2 3 1 0.15075060 1 0.15075060 0 1 6.508751956
#> 3 4 1 0.56212147 1 2.41915162 0 1 2.690053477
#> 4 6 1 0.47629626 1 0.47629626 0 1 0.024984612
#> 5 7 1 0.64391257 1 0.64391257 0 1 0.003990494
#> 6 9 1 0.27386179 1 0.27386179 0 1 5.236761118
#> 7 10 1 0.41250249 1 0.89799098 1 0 5.930290024
#> 8 12 1 0.62277501 1 0.64594684 0 1 4.695528368
#> 9 13 1 0.12997017 1 0.12997017 0 1 1.509620696
#> 10 14 1 1.04040922 1 1.04040922 0 1 1.242366128
#> 11 15 1 0.02544847 1 0.02544847 0 1 4.443782684
#> 12 17 1 0.57767544 1 0.57767544 0 1 0.823651729
#> 13 18 1 0.09734062 1 0.09734062 0 1 1.893173764
#> 14 19 1 0.16208977 1 0.16208977 0 1 4.184192105
#> 15 21 1 0.14169789 1 0.14169789 0 1 5.848798379
#> 16 22 1 0.04763364 1 0.04763364 0 1 3.433677910
#> 17 23 1 0.45706380 1 0.45706380 0 1 4.143891723
#> 18 24 1 0.09314261 1 0.91309530 0 1 2.018429318
#> 19 25 1 0.56389389 1 0.56389389 0 1 3.687071947
#> 20 26 1 0.47023947 1 0.47023947 0 1 4.227757026
#> 21 27 1 0.22703896 1 0.22703896 0 1 4.133027100
#> 22 29 1 0.46661062 1 0.51297590 0 1 6.289522646
#> 23 38 2 0.15540772 1 0.15540772 0 1 1.464038296
#> 24 40 2 0.04322971 1 0.04322971 1 0 6.785051292
#> 25 41 2 0.54570637 1 0.54570637 0 1 4.364613695
#> 26 43 2 0.24034500 0 0.24034500 1 0 6.587936003
#> 27 44 2 1.63214588 1 1.63214588 0 1 1.881478205
#> 28 45 2 0.58103357 1 0.58103357 0 1 5.467244969
#> 29 54 2 0.34380339 1 0.74697844 0 1 2.132945149
#> 30 59 2 0.09644431 1 0.27138395 0 1 5.442366586
#> 31 61 2 0.45262441 1 1.01968087 0 1 5.125696822
#> 32 62 2 0.05870165 1 0.05870165 0 1 5.439043818
#> 33 65 2 0.49377546 1 0.49377546 0 1 4.180423580
#> 34 68 2 0.12736172 1 0.12736172 0 1 3.157080468
#> 35 74 2 0.07764841 0 0.07764841 1 0 6.750632599
#> 36 76 2 0.60140979 1 0.60140979 0 1 5.403354042
#> 37 79 2 0.03248963 1 0.03248963 0 1 0.871058516
#> 38 82 2 0.19592203 1 0.19592203 0 1 0.017242287
#> 39 83 2 0.06006929 1 1.21931632 0 1 3.657871913
#> 40 86 2 0.11559171 1 0.11559171 0 1 3.960323525
#> 41 88 2 0.09813191 1 1.79531539 0 1 0.935697695
#> 42 89 2 0.51809902 1 0.51809902 0 1 1.062610177
#> OStimeCal PFStimeCal
#> 1 4.8874357 4.8874357
#> 2 6.6595026 6.6595026
#> 3 5.1092051 3.2521749
#> 4 0.5012809 0.5012809
#> 5 0.6479031 0.6479031
#> 6 5.5106229 5.5106229
#> 7 6.8282810 6.3427925
#> 8 5.3414752 5.3183034
#> 9 1.6395909 1.6395909
#> 10 2.2827753 2.2827753
#> 11 4.4692312 4.4692312
#> 12 1.4013272 1.4013272
#> 13 1.9905144 1.9905144
#> 14 4.3462819 4.3462819
#> 15 5.9904963 5.9904963
#> 16 3.4813115 3.4813115
#> 17 4.6009555 4.6009555
#> 18 2.9315246 2.1115719
#> 19 4.2509658 4.2509658
#> 20 4.6979965 4.6979965
#> 21 4.3600661 4.3600661
#> 22 6.8024985 6.7561333
#> 23 1.6194460 1.6194460
#> 24 6.8282810 6.8282810
#> 25 4.9103201 4.9103201
#> 26 6.8282810 6.8282810
#> 27 3.5136241 3.5136241
#> 28 6.0482785 6.0482785
#> 29 2.8799236 2.4767485
#> 30 5.7137505 5.5388109
#> 31 6.1453777 5.5783212
#> 32 5.4977455 5.4977455
#> 33 4.6741990 4.6741990
#> 34 3.2844422 3.2844422
#> 35 6.8282810 6.8282810
#> 36 6.0047638 6.0047638
#> 37 0.9035481 0.9035481
#> 38 0.2131643 0.2131643
#> 39 4.8771882 3.7179412
#> 40 4.0759152 4.0759152
#> 41 2.7310131 1.0338296
#> 42 1.5807092 1.5807092
simStudy_30POS
#> id trt PFStime PFSevent OStime CensoredOS OSevent recruitTime
#> 1 2 1 0.26116360 1 0.26116360 0 1 4.626272118
#> 2 4 1 0.56212147 1 2.41915162 0 1 2.690053477
#> 3 6 1 0.47629626 1 0.47629626 0 1 0.024984612
#> 4 7 1 0.64391257 1 0.64391257 0 1 0.003990494
#> 5 9 1 0.26098435 0 0.26098435 1 0 5.236761118
#> 6 12 1 0.62277501 1 0.64594684 0 1 4.695528368
#> 7 13 1 0.12997017 1 0.12997017 0 1 1.509620696
#> 8 14 1 1.04040922 1 1.04040922 0 1 1.242366128
#> 9 15 1 0.02544847 1 0.02544847 0 1 4.443782684
#> 10 17 1 0.57767544 1 0.57767544 0 1 0.823651729
#> 11 18 1 0.09734062 1 0.09734062 0 1 1.893173764
#> 12 19 1 0.16208977 1 0.16208977 0 1 4.184192105
#> 13 22 1 0.04763364 1 0.04763364 0 1 3.433677910
#> 14 23 1 0.45706380 1 0.45706380 0 1 4.143891723
#> 15 24 1 0.09314261 1 0.91309530 0 1 2.018429318
#> 16 25 1 0.56389389 1 0.56389389 0 1 3.687071947
#> 17 26 1 0.47023947 1 0.47023947 0 1 4.227757026
#> 18 27 1 0.22703896 1 0.22703896 0 1 4.133027100
#> 19 38 2 0.15540772 1 0.15540772 0 1 1.464038296
#> 20 41 2 0.54570637 1 0.54570637 0 1 4.364613695
#> 21 44 2 1.63214588 1 1.63214588 0 1 1.881478205
#> 22 45 2 0.03050050 0 0.03050050 1 0 5.467244969
#> 23 54 2 0.34380339 1 0.74697844 0 1 2.132945149
#> 24 59 2 0.05537888 0 0.05537888 1 0 5.442366586
#> 25 61 2 0.37204864 0 0.37204864 1 0 5.125696822
#> 26 62 2 0.05870165 1 0.05870165 0 1 5.439043818
#> 27 65 2 0.49377546 1 0.49377546 0 1 4.180423580
#> 28 68 2 0.12736172 1 0.12736172 0 1 3.157080468
#> 29 76 2 0.09439142 0 0.09439142 1 0 5.403354042
#> 30 79 2 0.03248963 1 0.03248963 0 1 0.871058516
#> 31 82 2 0.19592203 1 0.19592203 0 1 0.017242287
#> 32 83 2 0.06006929 1 1.21931632 0 1 3.657871913
#> 33 86 2 0.11559171 1 0.11559171 0 1 3.960323525
#> 34 88 2 0.09813191 1 1.79531539 0 1 0.935697695
#> 35 89 2 0.51809902 1 0.51809902 0 1 1.062610177
#> OStimeCal PFStimeCal
#> 1 4.8874357 4.8874357
#> 2 5.1092051 3.2521749
#> 3 0.5012809 0.5012809
#> 4 0.6479031 0.6479031
#> 5 5.4977455 5.4977455
#> 6 5.3414752 5.3183034
#> 7 1.6395909 1.6395909
#> 8 2.2827753 2.2827753
#> 9 4.4692312 4.4692312
#> 10 1.4013272 1.4013272
#> 11 1.9905144 1.9905144
#> 12 4.3462819 4.3462819
#> 13 3.4813115 3.4813115
#> 14 4.6009555 4.6009555
#> 15 2.9315246 2.1115719
#> 16 4.2509658 4.2509658
#> 17 4.6979965 4.6979965
#> 18 4.3600661 4.3600661
#> 19 1.6194460 1.6194460
#> 20 4.9103201 4.9103201
#> 21 3.5136241 3.5136241
#> 22 5.4977455 5.4977455
#> 23 2.8799236 2.4767485
#> 24 5.4977455 5.4977455
#> 25 5.4977455 5.4977455
#> 26 5.4977455 5.4977455
#> 27 4.6741990 4.6741990
#> 28 3.2844422 3.2844422
#> 29 5.4977455 5.4977455
#> 30 0.9035481 0.9035481
#> 31 0.2131643 0.2131643
#> 32 4.8771882 3.7179412
#> 33 4.0759152 4.0759152
#> 34 2.7310131 1.0338296
#> 35 1.5807092 1.5807092