Delayed Data Loading
Dawid Kałędkowski
15.05.2022
delayed-data-loading.RmdOverview
teal.data provides several ways to include datasets into
shiny applications. Normally, one develops an application in such a way
that the data is available before the app starts. This always involves
passing data.frame objects using the
cdisc_data or teal_data functions to the shiny
application. This way of building an app is applicable if data is
physically available before the app is created.
Including data to the shiny app as global objects means that they will be fixed and won’t change throughout the life of the application session.
The other possible scenario is that data may not be available during the creation or initialization of the application. Such data would need to be loaded by the shiny application after being initialized, which may also involve the entering of user name and password credentials via shiny UI components. Delayed data loading in applications involves specifying metadata which stores all information needed to pull the data from the right source. In delayed data loading applications, before the main app loads the user is prompted with data loading UI.
This means that delayed data applications can be two-staged:
- Data loading app
- Main app
Key definitions
The following are general descriptions for the main classes of the
teal.data package.
TealDatasetcontains physical data in the form of a singledata.frameplus reproducible code.TealDatasetConnectorcontains instructions to obtain a singleTealDatasetobject.TealDataConnectionopens and closes connections (with remote data sources).TealDataConnectorcontains arbitrary manyTealDatasetConnectorobjects and optionally oneTealDataConnectionobject.TealDatacontains arbitrary manyTealDataset,TealDatasetConnectorand / orTealDataConnectorobjects.
With the exception of TealDataConnection, all of the
above classes have their CDISC equivalent, e.g. TealData
-> CDISCTealData.
Creating an app
Callable
CallableFunction
This class won’t be used often by the developers but it’s essential
for all connectors to store and execute R functions with the feature to
get the code to reproduce the function calls. In the example below an
object of class CallableFunction is created which runs
synthetic_cdisc_dataset function with arguments specified
by the user.
library(teal.data)
#> Loading required package: shiny
library(scda)
#>
# initialize object
fun <- callable_function(fun = synthetic_cdisc_dataset)
# set arguments to function
fun$set_args(list(dataset_name = "adsl", name = "latest"))
# execute function with arguments set
df <- fun$run()
head(df, 3)
#> STUDYID USUBJID SUBJID SITEID AGE AGEU SEX
#> 1 AB12345 AB12345-CHN-3-id-128 id-128 CHN-3 32 YEARS M
#> 2 AB12345 AB12345-CHN-15-id-262 id-262 CHN-15 35 YEARS M
#> 3 AB12345 AB12345-RUS-3-id-378 id-378 RUS-3 30 YEARS F
#> RACE ETHNIC COUNTRY DTHFL INVID
#> 1 ASIAN NOT HISPANIC OR LATINO CHN N INV ID CHN-3
#> 2 BLACK OR AFRICAN AMERICAN NOT HISPANIC OR LATINO CHN N INV ID CHN-15
#> 3 ASIAN NOT HISPANIC OR LATINO RUS N INV ID RUS-3
#> INVNAM ARM ARMCD ACTARM ACTARMCD TRT01P
#> 1 Dr. CHN-3 Doe A: Drug X ARM A A: Drug X ARM A A: Drug X
#> 2 Dr. CHN-15 Doe C: Combination ARM C C: Combination ARM C C: Combination
#> 3 Dr. RUS-3 Doe C: Combination ARM C C: Combination ARM C C: Combination
#> TRT01A REGION1 STRATA1 STRATA2 BMRKR1 BMRKR2 ITTFL SAFFL BMEASIFL
#> 1 A: Drug X Asia C S2 14.424934 MEDIUM Y Y Y
#> 2 C: Combination Asia C S1 4.055463 LOW Y Y N
#> 3 C: Combination Eurasia A S1 2.803240 HIGH Y Y Y
#> BEP01FL RANDDT TRTSDTM TRTEDTM EOSSTT
#> 1 Y 2019-02-22 2019-02-24 11:09:18 2021-02-23 22:47:42 COMPLETED
#> 2 N 2019-02-26 2019-02-26 09:05:00 2021-02-25 20:43:24 COMPLETED
#> 3 N 2019-02-24 2019-02-28 03:19:08 2021-02-27 14:57:32 COMPLETED
#> EOTSTT EOSDT EOSDY DCSREAS DTHDT DTHCAUS DTHCAT LDDTHELD LDDTHGR1
#> 1 COMPLETED 2021-02-23 731 <NA> <NA> <NA> <NA> NA <NA>
#> 2 COMPLETED 2021-02-25 731 <NA> <NA> <NA> <NA> NA <NA>
#> 3 COMPLETED 2021-02-27 731 <NA> <NA> <NA> <NA> NA <NA>
#> LSTALVDT DTHADY study_duration_secs
#> 1 2021-03-05 NA 63113904
#> 2 2021-03-15 NA 63113904
#> 3 2021-03-15 NA 63113904
# check reproducible code
cat(fun$get_call())
#> scda::synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")It’s also possible to execute the fun$run() function
with arguments added on the fly in a named list. Dynamic
arguments won’t be reflected in the reproducible code, which will have
consequences in other places - we will get back to this in later stages
of the documentation.
# initialize object
fun <- callable_function(fun = synthetic_cdisc_dataset)
# add arguments on the fly
df <- fun$run(args = list(dataset_name = "adae", name = "latest"))
head(df, 3)
#> STUDYID USUBJID SUBJID SITEID AGE AGEU SEX RACE
#> 1 AB12345 AB12345-BRA-1-id-134 id-134 BRA-1 47 YEARS M WHITE
#> 2 AB12345 AB12345-BRA-1-id-134 id-134 BRA-1 47 YEARS M WHITE
#> 3 AB12345 AB12345-BRA-1-id-134 id-134 BRA-1 47 YEARS M WHITE
#> ETHNIC COUNTRY DTHFL INVID INVNAM ARM
#> 1 NOT HISPANIC OR LATINO BRA N INV ID BRA-1 Dr. BRA-1 Doe A: Drug X
#> 2 NOT HISPANIC OR LATINO BRA N INV ID BRA-1 Dr. BRA-1 Doe A: Drug X
#> 3 NOT HISPANIC OR LATINO BRA N INV ID BRA-1 Dr. BRA-1 Doe A: Drug X
#> ARMCD ACTARM ACTARMCD TRT01P TRT01A REGION1 STRATA1 STRATA2
#> 1 ARM A A: Drug X ARM A A: Drug X A: Drug X South America B S2
#> 2 ARM A A: Drug X ARM A A: Drug X A: Drug X South America B S2
#> 3 ARM A A: Drug X ARM A A: Drug X A: Drug X South America B S2
#> BMRKR1 BMRKR2 ITTFL SAFFL BMEASIFL BEP01FL RANDDT TRTSDTM
#> 1 6.462991 LOW Y Y Y N 2020-11-03 2020-11-04 03:50:33
#> 2 6.462991 LOW Y Y Y N 2020-11-03 2020-11-04 03:50:33
#> 3 6.462991 LOW Y Y Y N 2020-11-03 2020-11-04 03:50:33
#> TRTEDTM EOSSTT EOTSTT EOSDT EOSDY DCSREAS DTHDT
#> 1 2022-11-04 15:28:57 COMPLETED COMPLETED 2022-11-04 731 <NA> <NA>
#> 2 2022-11-04 15:28:57 COMPLETED COMPLETED 2022-11-04 731 <NA> <NA>
#> 3 2022-11-04 15:28:57 COMPLETED COMPLETED 2022-11-04 731 <NA> <NA>
#> DTHCAUS DTHCAT LDDTHELD LDDTHGR1 LSTALVDT DTHADY study_duration_secs ASEQ
#> 1 <NA> <NA> NA <NA> 2022-11-15 NA 63113904 1
#> 2 <NA> <NA> NA <NA> 2022-11-15 NA 63113904 2
#> 3 <NA> <NA> NA <NA> 2022-11-15 NA 63113904 3
#> AESEQ AETERM AELLT AEDECOD AEHLT AEHLGT
#> 1 1 trm B.2.1.2.1 llt B.2.1.2.1 dcd B.2.1.2.1 hlt B.2.1.2 hlgt B.2.1
#> 2 2 trm D.1.1.4.2 llt D.1.1.4.2 dcd D.1.1.4.2 hlt D.1.1.4 hlgt D.1.1
#> 3 3 trm A.1.1.1.2 llt A.1.1.1.2 dcd A.1.1.1.2 hlt A.1.1.1 hlgt A.1.1
#> AEBODSYS AESOC AESEV AESER AEACN AEREL
#> 1 cl B.2 cl B MODERATE N DOSE NOT CHANGED N
#> 2 cl D.1 cl D MODERATE N DOSE NOT CHANGED N
#> 3 cl A.1 cl A MODERATE Y DOSE NOT CHANGED N
#> AEOUT AESDTH TRTEMFL AECONTRT ASTDTM
#> 1 RECOVERED/RESOLVED N Y N 2021-07-13
#> 2 RECOVERING/RESOLVING N Y N 2021-09-04
#> 3 RECOVERED/RESOLVED WITH SEQUELAE N Y Y 2022-03-15
#> AENDTM ASTDY AENDY AETOXGR SMQ01NAM SMQ02NAM SMQ01SC SMQ02SC CQ01NAM
#> 1 2022-04-05 251 517 3 <NA> <NA> <NA> <NA> <NA>
#> 2 2022-05-16 304 558 3 <NA> <NA> <NA> <NA> <NA>
#> 3 2022-10-29 496 724 2 <NA> <NA> <NA> <NA> <NA>
#> ANL01FL AERELNST AEACNOTH
#> 1 Y OTHER PROCEDURE/SURGERY
#> 2 Y DISEASE UNDER STUDY MEDICATION
#> 3 Y DISEASE UNDER STUDY NONE
# dynamic arguments not reflected in the call
cat(fun$get_call())
#> scda::synthetic_cdisc_dataset()CallableFunction can also depend on other R objects as
the example function below depends on ADSL. The function
can be executed in specific environment where we can copy objects needed
to execute a call. To include objects in the function call, one has to
use $assign_to_env() to copy the object and
$set_args(list(ADSL = as.name(ADSL))) to link the object
with function argument.
adsl_raw <- synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")
fun <- callable_function(fun = function(adsl) {
adsl_2 <- adsl
adsl_2$new_col <- TRUE
adsl_2
})
# copy adsl to CallableFunction environment
fun$assign_to_env("adsl", adsl_raw)
# set arguments
fun$set_args(args = list(adsl = as.name("adsl")))
# execute function
df <- fun$run()
head(df, 3)
#> STUDYID USUBJID SUBJID SITEID AGE AGEU SEX
#> 1 AB12345 AB12345-CHN-3-id-128 id-128 CHN-3 32 YEARS M
#> 2 AB12345 AB12345-CHN-15-id-262 id-262 CHN-15 35 YEARS M
#> 3 AB12345 AB12345-RUS-3-id-378 id-378 RUS-3 30 YEARS F
#> RACE ETHNIC COUNTRY DTHFL INVID
#> 1 ASIAN NOT HISPANIC OR LATINO CHN N INV ID CHN-3
#> 2 BLACK OR AFRICAN AMERICAN NOT HISPANIC OR LATINO CHN N INV ID CHN-15
#> 3 ASIAN NOT HISPANIC OR LATINO RUS N INV ID RUS-3
#> INVNAM ARM ARMCD ACTARM ACTARMCD TRT01P
#> 1 Dr. CHN-3 Doe A: Drug X ARM A A: Drug X ARM A A: Drug X
#> 2 Dr. CHN-15 Doe C: Combination ARM C C: Combination ARM C C: Combination
#> 3 Dr. RUS-3 Doe C: Combination ARM C C: Combination ARM C C: Combination
#> TRT01A REGION1 STRATA1 STRATA2 BMRKR1 BMRKR2 ITTFL SAFFL BMEASIFL
#> 1 A: Drug X Asia C S2 14.424934 MEDIUM Y Y Y
#> 2 C: Combination Asia C S1 4.055463 LOW Y Y N
#> 3 C: Combination Eurasia A S1 2.803240 HIGH Y Y Y
#> BEP01FL RANDDT TRTSDTM TRTEDTM EOSSTT
#> 1 Y 2019-02-22 2019-02-24 11:09:18 2021-02-23 22:47:42 COMPLETED
#> 2 N 2019-02-26 2019-02-26 09:05:00 2021-02-25 20:43:24 COMPLETED
#> 3 N 2019-02-24 2019-02-28 03:19:08 2021-02-27 14:57:32 COMPLETED
#> EOTSTT EOSDT EOSDY DCSREAS DTHDT DTHCAUS DTHCAT LDDTHELD LDDTHGR1
#> 1 COMPLETED 2021-02-23 731 <NA> <NA> <NA> <NA> NA <NA>
#> 2 COMPLETED 2021-02-25 731 <NA> <NA> <NA> <NA> NA <NA>
#> 3 COMPLETED 2021-02-27 731 <NA> <NA> <NA> <NA> NA <NA>
#> LSTALVDT DTHADY study_duration_secs new_col
#> 1 2021-03-05 NA 63113904 TRUE
#> 2 2021-03-15 NA 63113904 TRUE
#> 3 2021-03-15 NA 63113904 TRUE
# get R code
fun$get_call()
#> [1] "(function(adsl) {\n adsl_2 <- adsl\n adsl_2$new_col <- TRUE\n adsl_2\n})(adsl = adsl)"
CallableCode
A simpler version of the Callable class is
CallableCode. Similar to CallableFunction,
CallableCode stores code which can be evaluated using
run() but it isn’t able to use dynamic arguments to make
the code more general. CallableCode also allows the
assignment of objects to its environment. CallableCode can
contain multiple lines (commands) of code and also allows
library calls. Please note that objects assigned to this
independent environment can’t be modified because they are locked
immediately. This means that the CallableCode created below
is not allowed to make any changes to x1.
code <- callable_code(
"library(scda)
ADTTE <- synthetic_cdisc_dataset(dataset_name = \"adtte\", name = \"latest\")
ADTTE$x1 <- x1
ADTTE <- dplyr::filter(ADTTE, PARAMCD %in% c('EFS', 'OS'))"
)
# examine call
cat(code$get_call())
#> library(scda)
#> ADTTE <- synthetic_cdisc_dataset(dataset_name = "adtte", name = "latest")
#> ADTTE$x1 <- x1
#> ADTTE <- dplyr::filter(ADTTE, PARAMCD %in% c("EFS", "OS"))
# assign x1 to environment (otherwise code would run with error as x1 would not be defined)
code$assign_to_env("x1", 1)
# evaluate call
df <- code$run()
head(df$x1, 3)
#> [1] 1 1 1
TealDataset (base class which
CDISCTealDataset inherits from)
TealDataset is an R6 class which keeps a
data.frame in its raw_data slot. One can
create a TealDataset by including data.frame
and setting data attributes. In the example below we first create
adsl and then put this data.frame into the
cdisc_dataset function. Together with
data.frame one should also provide a dataname
and optional code to reproduce the data if reproducibility
is required.
Note cdisc_dataset returns an object of
CDISCTealDataset, which is needed for CDISC analysis.
library(magrittr)
adsl_raw <- synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest") %>% head(3)
adsl_dataset <- cdisc_dataset(
dataname = "ADSL",
x = adsl_raw,
code = "ADSL <- synthetic_cdisc_dataset(dataset_name = \"adsl\", name = \"latest\") %>% head(3)"
)The object created above contains all previously defined attributes which can be extracted.
# check if code is reproducible
get_dataname(adsl_dataset)
#> [1] "ADSL"
# get label
get_dataset_label(adsl_dataset)
#> [1] "Subject Level Analysis Dataset"
# get reproducible code
get_code(adsl_dataset)
#> [1] "ADSL <- synthetic_cdisc_dataset(dataset_name = \"adsl\", name = \"latest\") %>% head(3)"
# get data.frame
get_raw_data(adsl_dataset)
#> # A tibble: 3 × 44
#> STUDYID USUBJID SUBJID SITEID AGE AGEU SEX RACE ETHNIC COUNTRY DTHFL
#> <chr> <chr> <chr> <chr> <int> <fct> <fct> <fct> <fct> <fct> <fct>
#> 1 AB12345 AB12345-CH… id-128 CHN-3 32 YEARS M ASIAN NOT H… CHN N
#> 2 AB12345 AB12345-CH… id-262 CHN-15 35 YEARS M BLAC… NOT H… CHN N
#> 3 AB12345 AB12345-RU… id-378 RUS-3 30 YEARS F ASIAN NOT H… RUS N
#> # … with 33 more variables: INVID <chr>, INVNAM <chr>, ARM <fct>, ARMCD <fct>,
#> # ACTARM <fct>, ACTARMCD <fct>, TRT01P <fct>, TRT01A <fct>, REGION1 <fct>,
#> # STRATA1 <fct>, STRATA2 <fct>, BMRKR1 <dbl>, BMRKR2 <fct>, ITTFL <fct>,
#> # SAFFL <fct>, BMEASIFL <fct>, BEP01FL <fct>, RANDDT <date>, TRTSDTM <dttm>,
#> # TRTEDTM <dttm>, EOSSTT <fct>, EOTSTT <fct>, EOSDT <date>, EOSDY <int>,
#> # DCSREAS <fct>, DTHDT <date>, DTHCAUS <fct>, DTHCAT <fct>, LDDTHELD <int>,
#> # LDDTHGR1 <fct>, LSTALVDT <date>, DTHADY <int>, study_duration_secs <dbl>
# get keys (i.e. the primary keys of the dataset)
adsl_dataset$get_keys()
#> [1] "STUDYID" "USUBJID"
TealDatasetConnector (base class which
CDISCTealDatasetConnector inherits from)
TealDatasetConnector contains a Callable
object to obtain a single TealDataset object. In the code
chunk below, a connector is created based on the
synthetic_cdisc_dataset function.
adsl_conn <- dataset_connector(
dataname = "ADSL",
pull_callable = callable_function("synthetic_cdisc_dataset") %>%
set_args(list(dataset_name = "adsl", name = "latest")),
keys = get_cdisc_keys("ADSL"),
label = "Subject-Level Analysis Dataset"
)Initially, adsl_conn doesn’t contain any data.
Attempting to fetch unavailable data will produce an error.
# get raw data
try(get_raw_data(adsl_conn))
#> Error : 'ADSL' has not been pulled yet
#> - please use `load_dataset()` first.Data can be loaded using load_dataset() function. Before
data is loaded, adsl_conn contains reproducible code which
is also the code to pull the data.
# execution/reproducible code
get_code(adsl_conn)
#> [1] "ADSL <- scda::synthetic_cdisc_dataset(dataset_name = \"adsl\", name = \"latest\")"
# pull data
load_dataset(adsl_conn)
# get raw data
get_raw_data(adsl_conn)
#> # A tibble: 400 × 44
#> STUDYID USUBJID SUBJID SITEID AGE AGEU SEX RACE ETHNIC COUNTRY DTHFL
#> <chr> <chr> <chr> <chr> <int> <fct> <fct> <fct> <fct> <fct> <fct>
#> 1 AB12345 AB12345-C… id-128 CHN-3 32 YEARS M ASIAN NOT H… CHN N
#> 2 AB12345 AB12345-C… id-262 CHN-15 35 YEARS M BLAC… NOT H… CHN N
#> 3 AB12345 AB12345-R… id-378 RUS-3 30 YEARS F ASIAN NOT H… RUS N
#> 4 AB12345 AB12345-C… id-220 CHN-11 26 YEARS F ASIAN NOT H… CHN N
#> 5 AB12345 AB12345-C… id-267 CHN-7 40 YEARS M ASIAN UNKNO… CHN N
#> 6 AB12345 AB12345-C… id-201 CHN-15 49 YEARS M ASIAN NOT H… CHN N
#> 7 AB12345 AB12345-U… id-45 USA-1 34 YEARS F ASIAN NOT H… USA N
#> 8 AB12345 AB12345-U… id-261 USA-1 32 YEARS F ASIAN NOT H… USA N
#> 9 AB12345 AB12345-N… id-173 NGA-11 24 YEARS F BLAC… NOT H… NGA N
#> 10 AB12345 AB12345-C… id-307 CHN-1 24 YEARS M ASIAN NOT H… CHN N
#> # … with 390 more rows, and 33 more variables: INVID <chr>, INVNAM <chr>,
#> # ARM <fct>, ARMCD <fct>, ACTARM <fct>, ACTARMCD <fct>, TRT01P <fct>,
#> # TRT01A <fct>, REGION1 <fct>, STRATA1 <fct>, STRATA2 <fct>, BMRKR1 <dbl>,
#> # BMRKR2 <fct>, ITTFL <fct>, SAFFL <fct>, BMEASIFL <fct>, BEP01FL <fct>,
#> # RANDDT <date>, TRTSDTM <dttm>, TRTEDTM <dttm>, EOSSTT <fct>, EOTSTT <fct>,
#> # EOSDT <date>, EOSDY <int>, DCSREAS <fct>, DTHDT <date>, DTHCAUS <fct>,
#> # DTHCAT <fct>, LDDTHELD <int>, LDDTHGR1 <fct>, LSTALVDT <date>, …TealDatasetConnector also allows other variables to be
passed to its Callable object that may depend on them. In
the code below the adsl_raw object - created above - is
added to the CallableFunction. To properly link the
ADSL argument of the function inside the
CallableFunction object with the raw data
adsl_raw several parameters have to match:
-
as.name("dummy_name")must be the value with name"ADSL"-set_args(list(ADSL = as.name("dummy_name"))) - and then
dummy_namemust be linked with the raw dataadsl_raw-vars = list(dummy_name = adsl_raw)
The name ADSL is fixed because it is the name of the
argument of the function inside of the CallableFunction.
The name dummy_name is free to be any valid R
name.
# here we use the general dataset_connector function which pulls an object of type TealDataset
# there is also a cdisc_dataset_connector function which pulls an object of type CDISCTealDataset
adsl_2 <- dataset_connector(
dataname = "ADSL_2",
pull_callable = callable_function(fun = function(ADSL) ADSL_2 <- ADSL) %>% # nolint
set_args(list(ADSL = as.name("dummy_name"))),
keys = get_cdisc_keys("ADSL"),
label = "Example label",
vars = list(dummy_name = adsl_raw)
)
load_dataset(adsl_2)TealDatasetConnector like the other delayed data objects
contains a launch method which can be used to obtain data
using shiny application. This function can be used to check if the
objects are specified correctly and to investigate potential mistakes.
By default the shiny app won’t render any inputs for datasets until we
specify them. To set inputs we should use $set_ui_input(),
by passing ui module function, with ns argument
(shiny namespace ID object). In the example below, the
callable_function object contains two arguments,
ADSL and n. ADSL is given while
n is entered from a shiny app after the launch
method is called.
adsl_3 <- dataset_connector(
dataname = "ADSL_3",
pull_callable = callable_function(fun = function(ADSL, n) ADSL_3 <- head(ADSL, n)) %>% # nolint
set_args(list(ADSL = as.name("ADSL"))),
keys = get_cdisc_keys("ADSL"),
label = "Example label",
vars = list(ADSL = adsl_raw)
)
adsl_3$set_ui_input(function(ns) {
list(
numericInput(inputId = ns("n"), label = "Choose number of records", min = 0, value = 1)
)
})
if (interactive()) {
adsl_3$launch()
}
TealDataConnection
Objects of this class are responsible to set a connection with remote
data sources. TealDataConnection opens and closes
connections by calling CallableFunction with the
appropriate arguments.
Note that if an app pulls data from a remote source, then the outputs will change even if the code is the same if the data in the remote source changes.
In shiny applications, connection arguments need to be
linked with inputs, which is why
TealDataConnection contains a module. Developers can
customize UI inputs to open the connection using
set_open_ui() and specify a relevant server function using
set_open_server(). It’s important to keep in mind that the
server module needs a connection as an argument to open()
and close() if needed.
open_fun <- callable_function(data.frame) # define opening function
open_fun$set_args(list(x = 1:5)) # define fixed arguments to opening function
close_fun <- callable_function(sum) # define closing function
close_fun$set_args(list(x = 1:5)) # define fixed arguments to closing function
ping_fun <- callable_function(function() TRUE)
x <- data_connection(
ping_fun = ping_fun, # define ping function
open_fun = open_fun, # define opening function
close_fun = close_fun # define closing function
)
TealDataConnector (base class which
CDISCTealDataConnector inherits from)
This class combines multiple TealDatasetConnector (or
CDISCTealDatasetConnector) and a single
TealDataConnection object. It creates a module to manage
connection and to load data. Below we create two
TealDatasetConnector objects and a
TealDataConnection object and we combine them together in a
TealDataConnector object.
# create TealDatasetConnectors
adsl <- dataset_connector(
dataname = "ADSL",
pull_callable = callable_function("synthetic_cdisc_dataset") %>%
set_args(list(dataset_name = "adsl", name = "latest")),
keys = get_cdisc_keys("ADSL"),
label = "Subject-Level Analysis Dataset"
)
adsl_3 <- dataset_connector(
dataname = "ADSL_3",
pull_callable = callable_function(fun = function(ADSL, name, n = 5) { # nolint
print(paste("slicing data from", name))
ADSL_3 <- head(ADSL, n) # nolint
}) %>%
set_args(list(ADSL = as.name("ADSL"))),
keys = get_cdisc_keys("ADSL"),
label = "Example label",
vars = list(ADSL = adsl)
)
adsl_3$set_ui_input(function(ns) {
list(
numericInput(inputId = ns("n"), label = "Choose number of records", min = 0, value = 1)
)
})
connectors <- list(adsl, adsl_3)
# create connection
scda_open_fun <- callable_function(fun = library)
scda_open_fun$set_args(list(package = "scda"))
scda_conn <- teal.data:::TealDataConnection$new(open_fun = scda_open_fun) # nolint
# create TealDataConnector
data <- teal.data:::TealDataConnector$new(
connection = scda_conn,
connectors = connectors
)The object created above can be used to pull data and obtain the
code. It combines code to pull the datasets preceded by any open
connection code and followed by any close connection code. Please note
that TealDataConnector doesn’t limit what kind of
connectors are set within, but one must be aware that it should rather
contain similar connectors (i.e. calling functions which share some
arguments). For example, if we execute
data$set_pull_args(args = list(name = "latest")), this
argument will be set for all connectors. In case when any connector
contains a CallableFunction which doesn’t have
name in its formals then it will fail.
CallableCode can’t hold any additional arguments as it’s
fixed and it will ignore every arguments set with
set_pull_args.
cat(get_code(data))
#> library(package = "scda")
#> ADSL <- scda::synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")
#> ADSL_3 <- (function(ADSL, name, n = 5) {
#> print(paste("slicing data from", name))
#> ADSL_3 <- head(ADSL, n)
#> })(ADSL = ADSL)
# pull ADSL and ADSL_3
data$set_pull_args(args = list(name = "latest"))
data$pull()
#> [1] "slicing data from latest"By default TealDataConnector creates simple shiny module
without any inputs to the arguments for the
callable_function of the connectors. This means that,
opening, closing, and pulling datasets are done on default arguments.
However, users can easily extend the module using set_ui()
and set_server() to specify the UI and server function
themselves. The general rule for creating these modules is that callback
from server to UI is not possible, because server module is executed
after the submit button is clicked. But see
set_preopen_server of TealDataConnection
object for a slight relaxation of this rule. Below we extend the app
interface by adding a text input which will be passed to the
scda function. The UI is created from
scda_conn and connectors object while the
server module requires connectors and
connection objects as additional arguments.
data$set_ui(
function(id, ...) {
ns <- NS(id)
tagList(
scda_conn$get_open_ui(ns("open_connection")),
textInput(ns("name"), p("Choose", code("name")), value = "latest"),
do.call(
what = "tagList",
args = lapply(
connectors,
function(connector) {
div(
connector$get_ui(
id = ns(connector$get_dataname())
),
br()
)
}
)
)
)
}
)
data$set_server(
function(id, connection, connectors) {
moduleServer(
id = id,
module = function(input, output, session) {
# opens connection
if (!is.null(connection$get_open_server())) {
connection$get_open_server()(
id = "open_connection",
connection = connection
)
}
for (connector in connectors) {
# set_args before to return them in the code (fixed args)
set_args(connector, args = list(name = input$name))
# pull each dataset
connector$get_server()(id = connector$get_dataname())
if (connector$is_failed()) {
break
}
}
}
)
}
)Executing data$launch() will open a shiny application
that prompts the user for input to load the data. Remember that data can
be loaded only once. So if you run the code to data$pull()
above please reinitialize the connectors, connection and data object
again before running the code below.
if (interactive()) {
data$launch()
}
TealData (base class which CDISCTealData
inherits from)
TealData manages TealDataset,
TealDatasetConnector and / or
TealDataConnector objects. CDISCTealData is
the equivalent object when creating apps to analyze CDISC data. These
objects are created using teal_data and
cdisc_data respectively. When using the teal
package these objects are passed as the data argument into
init.
data <- cdisc_data(
cdisc_dataset(
dataname = "ADSL",
synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest"),
code = "ADSL <- synthetic_cdisc_dataset(dataset_name = \"adsl\", name = \"latest\")"
),
cdisc_dataset(
dataname = "ADTTE",
synthetic_cdisc_dataset(dataset_name = "adtte", name = "latest"),
code = "ADTTE <- synthetic_cdisc_dataset(dataset_name = \"adtte\", name = \"latest\")"
)
)One can combine multiple delayed data objects using
[teal|cdisc]_data functions and include them in a shiny
application. TealData gathers all objects and sets combined
UI with shiny inputs from all objects. In the code chunk below we
specified four dataset connectors, but it works with arbitrary many
objects of any combination of the classes TealDataset,
TealDatasetConnector and / or
TealDataConnector.
adsl <- scda_cdisc_dataset_connector("ADSL", "adsl")
adae <- scda_cdisc_dataset_connector("ADAE", "adae")
advs <- scda_cdisc_dataset_connector("ADVS", "advs")
adtte <- scda_cdisc_dataset_connector("ADTTE", "adtte")
data <- cdisc_data(adsl, adae, advs, adtte)TealData also contains a launch() method to
investigate if data is set correctly. However, TealData
lacks the pull() method.
if (interactive()) {
data$launch()
}Reproducible code attached to the data object is
combined code of all components, but one can also extract code from
single dataset by specifying dataname argument.
get_code(data)
#> [1] "ADSL <- scda::synthetic_cdisc_dataset(dataset_name = \"adsl\", name = \"latest\")\nADAE <- scda::synthetic_cdisc_dataset(dataset_name = \"adae\", name = \"latest\")\nADVS <- scda::synthetic_cdisc_dataset(dataset_name = \"advs\", name = \"latest\")\nADTTE <- scda::synthetic_cdisc_dataset(dataset_name = \"adtte\", name = \"latest\")"
get_code(data, dataname = "ADSL")
#> [1] "ADSL <- scda::synthetic_cdisc_dataset(dataset_name = \"adsl\", name = \"latest\")"
get_code(data, dataname = "ADTTE")
#> [1] "ADTTE <- scda::synthetic_cdisc_dataset(dataset_name = \"adtte\", name = \"latest\")"Developers can also extract datasets and connectors included in the
TealData.
# if you launched the shiny app and pressed the submit button above to load the data,
# then these 4 lines do not need to be run
adsl$pull()
adae$pull()
advs$pull()
adtte$pull()
# get loaded datasets
data$get_datasets()
# get single dataset
data$get_dataset(dataname = "ADSL")
# get data and dataset connectors
data$get_connectors()
# get all datasets/connectors
data$get_items()Data modification
To modify a single dataset one should use mutate_dataset
by specifying code argument with code as a single character
or script with location of the script file. In the case of
delayed data, the pre-processing code passed into
mutate_dataset would be run after the data becomes
available.
adsl <- cdisc_dataset(
dataname = "ADSL",
synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest"),
code = "ADSL <- synthetic_cdisc_dataset(dataset_name = \"adsl\", name = \"latest\")"
) %>%
mutate_dataset(code = "ADSL$x1 <- 1")
adtte <- scda_cdisc_dataset_connector(
dataname = "ADTTE", "adtte"
) %>%
mutate_dataset(code = "ADTTE$x2 <- 2")get_code() function returns loading code from
CallableFunction and mutation code as provided in using
mutate_dataset.
cat(get_code(adsl))
#> ADSL <- synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")
#> ADSL$x1 <- 1
cat(get_code(adtte))
#> ADTTE <- scda::synthetic_cdisc_dataset(dataset_name = "adtte", name = "latest")
#> ADTTE$x2 <- 2mutate_dataset can also be used on TealData
objects which applies the code to the dataset specified in
dataname. This is important, as TealData can
track each call which affects this particular dataset. Check the code
below, where we create data containing two datasets using the
cdisc_data function. We can still use
mutate_dataset on the object which contains multiple
datasets, with one requirement - one needs to specify
dataname. Afterwards, one can extract reproducible code
from data which affects this particular dataname.
data <- cdisc_data(adsl, adtte) %>%
mutate_dataset(code = "ADSL$x3 <- 3", dataname = "ADSL")
# get reproducible code of all datasets
cat(get_code(data))
#> ADSL <- synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")
#> ADSL$x1 <- 1
#> ADTTE <- scda::synthetic_cdisc_dataset(dataset_name = "adtte", name = "latest")
#> ADTTE$x2 <- 2
#> ADSL$x3 <- 3
# get ADSL reproducible code
cat(get_code(data, dataname = "ADSL"))
#> ADSL <- synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")
#> ADSL$x1 <- 1
#> ADSL$x3 <- 3One can also pipe multiple mutate_dataset calls applied
on different datasets.
data <- mutate_dataset(data, code = "ADTTE$x4 <- 4", dataname = "ADTTE") %>%
mutate_dataset(code = "ADSL <- dplyr::filter(ADSL, SEX == 'F')", dataname = "ADSL")
cat(get_code(data, "ADSL"))
#> ADSL <- synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")
#> ADSL$x1 <- 1
#> ADSL$x3 <- 3
#> ADSL <- dplyr::filter(ADSL, SEX == "F")
# note that the code for ADTTE below does not contain any mention of ADSL
cat(get_code(data, "ADTTE"))
#> ADTTE <- scda::synthetic_cdisc_dataset(dataset_name = "adtte", name = "latest")
#> ADTTE$x2 <- 2
#> ADTTE$x4 <- 4Sometimes code of one object can depend on another, in this case we
can link them in the same way we link TealDatasetConnector
objects. If object code is dependent on another, as ADTTE depends on
ADSL below, get_code will return everything we need to be
executed to reproduce this dataset.
data <- mutate_dataset(
data,
code = "ADTTE <- filter(ADTTE, USUBJID %in% ADSL$USUBJID)",
dataname = "ADTTE",
vars = list(ADSL = adsl) # vars = list(<DATANAME> = <dataset name>))
) %>%
mutate_dataset("ADSL$var_created_after <- NA", dataname = "ADSL")
# note that the code that defines ADSL is now part of the code of ADTTE below
cat(get_code(data, dataname = "ADTTE"))
#> ADSL <- synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")
#> ADSL$x1 <- 1
#> ADTTE <- scda::synthetic_cdisc_dataset(dataset_name = "adtte", name = "latest")
#> ADTTE$x2 <- 2
#> ADSL$x3 <- 3
#> ADTTE$x4 <- 4
#> ADSL <- dplyr::filter(ADSL, SEX == "F")
#> ADTTE <- filter(ADTTE, USUBJID %in% ADSL$USUBJID)
# moreover, note that the code inserted to the mutate_dataset after the pipe was not included.Using mutate_dataset creates a tree of calls, which can
be subset by dataname, but developers can also use
mutate_data function which doesn’t require
dataname to be specified. When mutate_data is
used, code for a single dataset is not possible to be subset, instead
code of all datasets is returned.
data <- mutate_data(data,
code = "
ADSL$x3 <- 3
proxy_var <- 4
ADTTE$x4 <- proxy_var
"
)
# single dataset code is not possible to obtain anymore
cat(adsl_code <- get_code(data, "ADSL"))
#> ADSL <- synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")
#> ADSL$x1 <- 1
#> ADTTE <- scda::synthetic_cdisc_dataset(dataset_name = "adtte", name = "latest")
#> ADTTE$x2 <- 2
#> ADSL$x3 <- 3
#> ADTTE$x4 <- 4
#> ADSL <- dplyr::filter(ADSL, SEX == "F")
#> ADTTE <- filter(ADTTE, USUBJID %in% ADSL$USUBJID)
#> ADSL$var_created_after <- NA
#> ADSL$x3 <- 3
#> proxy_var <- 4
#> ADTTE$x4 <- proxy_var
cat(adtte_code <- get_code(data, "ADTTE"))
#> ADSL <- synthetic_cdisc_dataset(dataset_name = "adsl", name = "latest")
#> ADSL$x1 <- 1
#> ADTTE <- scda::synthetic_cdisc_dataset(dataset_name = "adtte", name = "latest")
#> ADTTE$x2 <- 2
#> ADSL$x3 <- 3
#> ADTTE$x4 <- 4
#> ADSL <- dplyr::filter(ADSL, SEX == "F")
#> ADTTE <- filter(ADTTE, USUBJID %in% ADSL$USUBJID)
#> ADSL$var_created_after <- NA
#> ADSL$x3 <- 3
#> proxy_var <- 4
#> ADTTE$x4 <- proxy_var
# TRUE
as.character(adsl_code) == as.character(adtte_code)
#> [1] TRUE
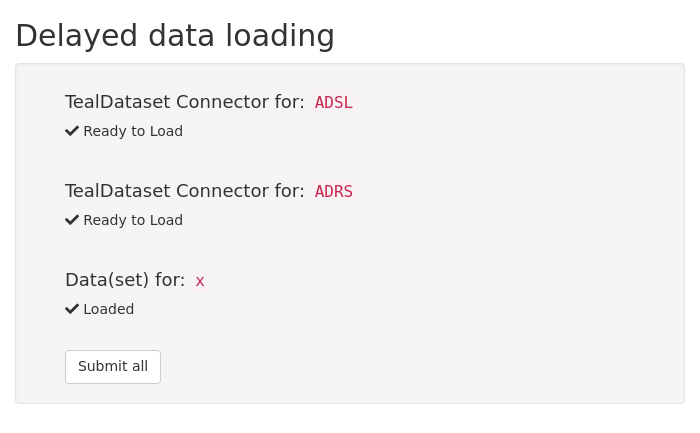
teal.data in a shiny app
adsl <- scda_cdisc_dataset_connector("ADSL", "adsl")
adrs <- scda_cdisc_dataset_connector("ADRS", "adrs")
x <- dataset("x", data.frame(x = 1, b = 2), code = "x <- data.frame(x = 1, b = 2)")
data <- teal_data(adsl, adrs, x)
shinyApp(
ui = fluidPage(
shinyjs::useShinyjs(),
titlePanel("Delayed data loading"),
sidebarLayout(
sidebarPanel(data$get_ui("data"), uiOutput("dataset")),
mainPanel(tableOutput("dist_plot"))
)
),
server = function(input, output, session) {
data_reactive <- data$get_server()("data")
observeEvent(data_reactive(), ignoreNULL = TRUE, {
shinyjs::hide("data-delayed_data")
})
output$dataset <- renderUI({
req(data_reactive())
datanames <- names(get_raw_data(data_reactive()))
radioButtons("dataname", "Select dataname", datanames, datanames[1])
})
output$dist_plot <- renderTable({
req(input$dataname)
dataset <- get_raw_data(data_reactive())[[input$dataname]]
head(dataset)
})
}
)
#>
#> Listening on http://127.0.0.1:4869