Introduction to {rtables}
Gabriel Becker and Adrian Waddell
2026-01-03
Source:vignettes/rtables.Rmd
rtables.RmdIntroduction
The rtables package provides a framework to create,
tabulate, and output tables in R. Most of the design requirements for
rtables have their origin in studying tables that are
commonly used to report analyses from clinical trials; however, we were
careful to keep rtables a general purpose toolkit.
In this vignette, we give a short introduction into
rtables and tabulating a table.
The content in this vignette is based on the following two resources:
- The
rtablesuseR 2020 presentation by Gabriel Becker -
rtables- A Framework For Creating Complex Structured Reporting Tables Via Multi-Level Faceted Computations.
The packages used in this vignette are rtables and
dplyr:
Overview
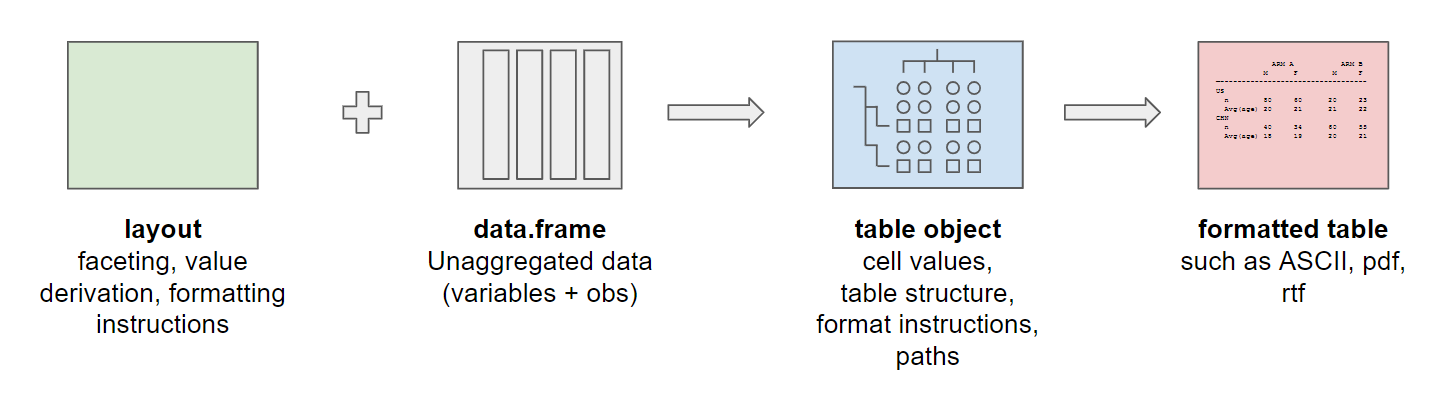
To build a table using rtables two components are
required: A layout constructed using rtables functions, and
a data.frame of unaggregated data. These two elements are
combined to build a table object. Table objects contain information
about both the content and the structure of the table, as well as
instructions on how this information should be processed to construct
the table. After obtaining the table object, a formatted table can be
printed in ASCII format, or exported to a variety of other formats
(.txt, .pdf, .docx, etc.).
Data
The data used in this vignette is a made up using random number generators. The data content is relatively simple: one row per imaginary person and one column per measurement: study arm, the country of origin, gender, handedness, age, and weight.
n <- 400
set.seed(1)
df <- tibble(
arm = factor(sample(c("Arm A", "Arm B"), n, replace = TRUE), levels = c("Arm A", "Arm B")),
country = factor(sample(c("CAN", "USA"), n, replace = TRUE, prob = c(.55, .45)), levels = c("CAN", "USA")),
gender = factor(sample(c("Female", "Male"), n, replace = TRUE), levels = c("Female", "Male")),
handed = factor(sample(c("Left", "Right"), n, prob = c(.6, .4), replace = TRUE), levels = c("Left", "Right")),
age = rchisq(n, 30) + 10
) %>% mutate(
weight = 35 * rnorm(n, sd = .5) + ifelse(gender == "Female", 140, 180)
)
head(df)
# # A tibble: 6 × 6
# arm country gender handed age weight
# <fct> <fct> <fct> <fct> <dbl> <dbl>
# 1 Arm A USA Female Left 31.3 139.
# 2 Arm B CAN Female Right 50.5 116.
# 3 Arm A USA Male Right 32.4 186.
# 4 Arm A USA Male Right 34.6 169.
# 5 Arm B USA Female Right 43.0 160.
# 6 Arm A USA Female Right 43.2 126.Note that we use factor variables so that the level order is
represented in the row or column order when we tabulate the information
of df below.
Building a Table
The aim of this vignette is to build the following table step by step:
# Arm A Arm B
# Female Male Female Male
# (N=96) (N=105) (N=92) (N=107)
# ————————————————————————————————————————————————————————————
# CAN 45 (46.9%) 64 (61.0%) 46 (50.0%) 62 (57.9%)
# Left 32 (33.3%) 42 (40.0%) 26 (28.3%) 37 (34.6%)
# mean 38.87 40.43 40.33 37.68
# Right 13 (13.5%) 22 (21.0%) 20 (21.7%) 25 (23.4%)
# mean 36.64 40.19 40.16 40.65
# USA 51 (53.1%) 41 (39.0%) 46 (50.0%) 45 (42.1%)
# Left 34 (35.4%) 19 (18.1%) 25 (27.2%) 25 (23.4%)
# mean 40.36 39.68 39.21 40.07
# Right 17 (17.7%) 22 (21.0%) 21 (22.8%) 20 (18.7%)
# mean 36.94 39.80 38.53 39.02Quick Start
The table above can be achieved via the qtable()
function. If you are new to tabulation with the rtables
layout framework, you can use this convenience wrapper to create many
types of two-way frequency tables.
The purpose of qtable is to enable quick exploratory
data analysis. See the exploratory_analysis
vignette for more details.
Here is the code to recreate the table above:
qtable(df,
row_vars = c("country", "handed"),
col_vars = c("arm", "gender"),
avar = "age",
afun = mean,
summarize_groups = TRUE,
row_labels = "mean"
)
# Arm A Arm B
# Female Male Female Male
# age - mean (N=96) (N=105) (N=92) (N=107)
# ——————————————————————————————————————————————————————————————
# CAN 45 (46.9%) 64 (61.0%) 46 (50.0%) 62 (57.9%)
# Left 32 (33.3%) 42 (40.0%) 26 (28.3%) 37 (34.6%)
# mean 38.87 40.43 40.33 37.68
# Right 13 (13.5%) 22 (21.0%) 20 (21.7%) 25 (23.4%)
# mean 36.64 40.19 40.16 40.65
# USA 51 (53.1%) 41 (39.0%) 46 (50.0%) 45 (42.1%)
# Left 34 (35.4%) 19 (18.1%) 25 (27.2%) 25 (23.4%)
# mean 40.36 39.68 39.21 40.07
# Right 17 (17.7%) 22 (21.0%) 21 (22.8%) 20 (18.7%)
# mean 36.94 39.80 38.53 39.02From the qtable function arguments above we can see many
of the key concepts of the underlying rtables layout
framework. The user needs to define:
- Which variables should be used as facets in the row and/or column space?
- Which variable should be used in the summary analysis?
- Which function should be used as a summary?
- Should the table include any marginal summaries?
- Are any labels needed to clarify the table content?
In the sections below we will look at translating each of these
questions to a set of features part of the rtables layout
framework. Now let’s take a look at building the example table with a
layout.
Layout Instructions
In rtables a basic table is defined to have 0 rows and
one column representing all data. Analyzing a variable is one way of
adding a row:
lyt <- basic_table() %>%
analyze("age", mean, format = "xx.x")
tbl <- build_table(lyt, df)
tbl
# all obs
# ——————————————
# mean 39.4In the code above we first described the table and assigned that
description to a variable lyt. We then built the table
using the actual data with build_table(). The description
of a table is called a table layout. basic_table() is the
start of every table layout and contains the information that we have in
one column representing all data. The analyze() instruction
adds to the layout that the age variable should be analyzed
with the mean() analysis function and the result should be
rounded to 1 decimal place.
Hence, a layout is “pre-data”, that is, it’s a description of how to build a table once we get data. We can look at the layout isolated:
lyt
# A Pre-data Table Layout
#
# Column-Split Structure:
# ()
#
# Row-Split Structure:
# age (** analysis **)The general layouting instructions are summarized below:
-
basic_table()is a layout representing a table with zero rows and one column - Nested splitting
- Summarizing Groups:
summarize_row_groups() - Analyzing Variables:
analyze(),analyze_colvars()
Using those functions, it is possible to create a wide variety of tables as we will show in this document.
Adding Column Structure
We will now add more structure to the columns by adding a column
split based on the factor variable arm:
lyt <- basic_table() %>%
split_cols_by("arm") %>%
analyze("age", afun = mean, format = "xx.x")
tbl <- build_table(lyt, df)
tbl
# Arm A Arm B
# ————————————————————
# mean 39.5 39.4The resulting table has one column per factor level of
arm. So the data represented by the first column is
df[df$arm == "ARM A", ]. Hence, the
split_cols_by() partitions the data among the columns by
default.
Column splitting can be done in a recursive/nested manner by adding
sequential split_cols_by() layout instruction. It’s also
possible to add a non-nested split. Here we splitting each arm further
by the gender:
lyt <- basic_table() %>%
split_cols_by("arm") %>%
split_cols_by("gender") %>%
analyze("age", afun = mean, format = "xx.x")
tbl <- build_table(lyt, df)
tbl
# Arm A Arm B
# Female Male Female Male
# ————————————————————————————————————
# mean 38.8 40.1 39.6 39.2The first column represents the data in df where
df$arm == "A" & df$gender == "Female" and the second
column the data in df where
df$arm == "A" & df$gender == "Male", and so on.
More information on column structure can be found in the col_counts
vignette.
Adding Row Structure
So far, we have created layouts with analysis and column splitting
instructions, i.e. analyze() and
split_cols_by(), respectively. This resulted with a table
with multiple columns and one data row. We will add more row structure
by stratifying the mean analysis by country (i.e. adding a split in the
row space):
lyt <- basic_table() %>%
split_cols_by("arm") %>%
split_cols_by("gender") %>%
split_rows_by("country") %>%
analyze("age", afun = mean, format = "xx.x")
tbl <- build_table(lyt, df)
tbl
# Arm A Arm B
# Female Male Female Male
# ——————————————————————————————————————
# CAN
# mean 38.2 40.3 40.3 38.9
# USA
# mean 39.2 39.7 38.9 39.6In this table the data used to derive the first data cell (average of
age of female Canadians in Arm A) is where
df$country == "CAN" & df$arm == "Arm A" & df$gender == "Female".
This cell value can also be calculated manually:
mean(df$age[df$country == "CAN" & df$arm == "Arm A" & df$gender == "Female"])
# [1] 38.22447Row structure can also be used to group the table into titled groups
of pages during rendering. We do this via ‘page by splits’, which are
declared via page_by = TRUE within a call to
split_rows_by:
lyt <- basic_table() %>%
split_cols_by("arm") %>%
split_cols_by("gender") %>%
split_rows_by("country", page_by = TRUE) %>%
split_rows_by("handed") %>%
analyze("age", afun = mean, format = "xx.x")
tbl <- build_table(lyt, df)
cat(export_as_txt(tbl, page_type = "letter", page_break = "\n\n~~~~~~ Page Break ~~~~~~\n\n"))
#
# country: CAN
#
# ————————————————————————————————————————
# Arm A Arm B
# Female Male Female Male
# ————————————————————————————————————————
# Left
# mean 38.9 40.4 40.3 37.7
# Right
# mean 36.6 40.2 40.2 40.6
#
#
# ~~~~~~ Page Break ~~~~~~
#
#
# country: USA
#
# ————————————————————————————————————————
# Arm A Arm B
# Female Male Female Male
# ————————————————————————————————————————
# Left
# mean 40.4 39.7 39.2 40.1
# Right
# mean 36.9 39.8 38.5 39.0We go into more detail on page-by splits and how to control the page-group specific titles in the Title and footer vignette.
Note that if you print or render a table without pagination, the page_by splits are currently rendered as normal row splits. This may change in future releases.
Adding Group Information
When adding row splits, we get by default label rows for each split
level, for example CAN and USA in the table
above. Besides the column space subsetting, we have now further
subsetted the data for each cell. It is often useful when defining a row
splitting to display information about each row group. In
rtables this is referred to as content information,
i.e. mean() on row 2 is a descendant of CAN
(visible via the indenting, though the table has an underlying tree
structure that is not of importance for this vignette). In order to add
content information and turn the CAN label row into a
content row, the summarize_row_groups() function is
required. By default, the count (nrows()) and percentage of
data relative to the column associated data is calculated:
lyt <- basic_table() %>%
split_cols_by("arm") %>%
split_cols_by("gender") %>%
split_rows_by("country") %>%
summarize_row_groups() %>%
analyze("age", afun = mean, format = "xx.x")
tbl <- build_table(lyt, df)
tbl
# Arm A Arm B
# Female Male Female Male
# ——————————————————————————————————————————————————————————
# CAN 45 (46.9%) 64 (61.0%) 46 (50.0%) 62 (57.9%)
# mean 38.2 40.3 40.3 38.9
# USA 51 (53.1%) 41 (39.0%) 46 (50.0%) 45 (42.1%)
# mean 39.2 39.7 38.9 39.6The relative percentage for average age of female Canadians is calculated as follows:
df_cell <- subset(df, df$country == "CAN" & df$arm == "Arm A" & df$gender == "Female")
df_col_1 <- subset(df, df$arm == "Arm A" & df$gender == "Female")
c(count = nrow(df_cell), percentage = nrow(df_cell) / nrow(df_col_1))
# count percentage
# 45.00000 0.46875so the group percentages per row split sum up to 1 for each column.
We can further split the row space by dividing each country by handedness:
lyt <- basic_table() %>%
split_cols_by("arm") %>%
split_cols_by("gender") %>%
split_rows_by("country") %>%
summarize_row_groups() %>%
split_rows_by("handed") %>%
analyze("age", afun = mean, format = "xx.x")
tbl <- build_table(lyt, df)
tbl
# Arm A Arm B
# Female Male Female Male
# ————————————————————————————————————————————————————————————
# CAN 45 (46.9%) 64 (61.0%) 46 (50.0%) 62 (57.9%)
# Left
# mean 38.9 40.4 40.3 37.7
# Right
# mean 36.6 40.2 40.2 40.6
# USA 51 (53.1%) 41 (39.0%) 46 (50.0%) 45 (42.1%)
# Left
# mean 40.4 39.7 39.2 40.1
# Right
# mean 36.9 39.8 38.5 39.0Next, we further add a count and percentage summary for handedness within each country:
lyt <- basic_table() %>%
split_cols_by("arm") %>%
split_cols_by("gender") %>%
split_rows_by("country") %>%
summarize_row_groups() %>%
split_rows_by("handed") %>%
summarize_row_groups() %>%
analyze("age", afun = mean, format = "xx.x")
tbl <- build_table(lyt, df)
tbl
# Arm A Arm B
# Female Male Female Male
# ————————————————————————————————————————————————————————————
# CAN 45 (46.9%) 64 (61.0%) 46 (50.0%) 62 (57.9%)
# Left 32 (33.3%) 42 (40.0%) 26 (28.3%) 37 (34.6%)
# mean 38.9 40.4 40.3 37.7
# Right 13 (13.5%) 22 (21.0%) 20 (21.7%) 25 (23.4%)
# mean 36.6 40.2 40.2 40.6
# USA 51 (53.1%) 41 (39.0%) 46 (50.0%) 45 (42.1%)
# Left 34 (35.4%) 19 (18.1%) 25 (27.2%) 25 (23.4%)
# mean 40.4 39.7 39.2 40.1
# Right 17 (17.7%) 22 (21.0%) 21 (22.8%) 20 (18.7%)
# mean 36.9 39.8 38.5 39.0Comparing with Other Tabulation Frameworks
There are a number of other table frameworks available in
R, including:
There are a number of reasons to choose rtables (yet
another tables R package):
- Output tables in ASCII to text files.
- Table rendering (ASCII, HTML, etc.) is separate from the data model. Hence, one always has access to the non-rounded/non-formatted numbers.
- Pagination in both horizontal and vertical directions to meet the health authority submission requirements.
- Cell, row, column, and table reference system.
- Titles, footers, and referential footnotes.
- Path based access to cell content which is useful for automated content generation.
More in depth comparisons of the various tabulation frameworks can be found in the Overview of table R packages chapter of the Tables in Clinical Trials with R book compiled by the R Consortium Tables Working Group.
Summary
In this vignette you have learned:
- Every cell has an associated subset of data - this means that much of tabulation has to do with splitting/subsetting data.
- Tables can be described with pre-data using layouts.
- Tables are a form of visualization of data.
The other vignettes in the rtables package will provide
more detailed information about the rtables package. We
recommend that you continue with the tabulation_dplyr
vignette which compares the information derived by the table in this
vignette using dplyr.