GENERAL
General Concepts
chevron is a collection of functions to creates tables,
listings, and graphs following Roche standards for clinical trials
reporting. After loading the R packages and the trial data, the output
is to be created by the main function run(...) . Two
arguments object= and adam_db= are always
expected in the function. object= specifies which Roche
Standard Template ID to use. adam_db= specifies the input
dataset. Other mandatory and optional arguments within the
run function vary depending on which template ID is called.
To access which arguments are required and what functions are used in
each template, simply try ?template
(e.g. ?aet01) to see more detailed descriptions and
instructions.
1. Input dataset and dataset names
The input dataset expected by the argument adam_db= in
the run(...) function is a collection of ADaM
datasets as a list object. Each ADaM dataset is expected to
be an object of data frame. If the ADaM datasets are read
in individually, user will need to combine them into a list object and
provide the name of the list to adam_db=. Also, each
element in the list are expected to have corresponding ADaM
dataset names. Conventional ADaM dataset names, including
adsl,adex, adae,
adlb,advs,adeg,adcm,admh,adrs,
and adtte, can be picked up by chevron with
one exception.
2. Expected variables in input analysis dataset
By default, chevron does not pull any subject-level
information from either adsl or adsub and
merge into the analysis dataset in the underlying preprocessing steps.
The analysis dataset fed into adam_db= is expected to have
all variables required for analysis available.
3. Character vs Factor
In the output generation, we often need to specify a particular
sorting order of a variable at the time of display. In
chevron, a character variable needs to be factorized with
pre-specified levels to display in order. When encountering cases, for
instance, "ARM A" has an Asian group only while
"ARM B" has both Asian and White groups, it is not able to
produce outputs like the demographic table unless "RACE" is
factorized to provide access to the same level attribute of the variable
"RACE" after the arm split. It is noted that the feature
comes from rtables instead of chevron.
proc_data <- syn_data
proc_data$adsl <- proc_data$adsl %>%
mutate(RACE = case_when(
ARMCD == "ARM A" ~ "ASIAN",
ARMCD == "ARM B" & !.data$RACE %in% c("WHITE", "ASIAN") ~ "ASIAN",
TRUE ~ RACE
))Having "RACE" as a character variable rather than a
factor leads to error message showing up as “Error: Error applying
analysis function (var - RACE): Number of rows generated by analysis
function do not match across all columns,” and it is recommended to
convert analysis variable "RACE" to a factor.
run(dmt01, proc_data)To resolve this issue, simply try factorizing the variable
"RACE":
proc_data$adsl$RACE <- as.factor(proc_data$adsl$RACE)
run(dmt01, proc_data)
#> A: Drug X B: Placebo C: Combination All Patients
#> (N=15) (N=15) (N=15) (N=45)
#> ————————————————————————————————————————————————————————————————————————————————————————————
#> Age (yr)
#> n 15 15 15 45
#> Mean (SD) 31.3 (5.3) 35.1 (9.0) 36.6 (6.4) 34.3 (7.3)
#> Median 31.0 35.0 35.0 34.0
#> Min - Max 24 - 40 24 - 57 24 - 49 24 - 57
#> Age Group
#> n 15 15 15 45
#> <65 15 (100%) 15 (100%) 15 (100%) 45 (100%)
#> Sex
#> n 15 15 15 45
#> Male 3 (20.0%) 7 (46.7%) 5 (33.3%) 15 (33.3%)
#> Female 12 (80.0%) 8 (53.3%) 10 (66.7%) 30 (66.7%)
#> Ethnicity
#> n 15 15 15 45
#> HISPANIC OR LATINO 2 (13.3%) 0 0 2 (4.4%)
#> NOT HISPANIC OR LATINO 13 (86.7%) 15 (100%) 13 (86.7%) 41 (91.1%)
#> NOT REPORTED 0 0 2 (13.3%) 2 (4.4%)
#> RACE
#> n 15 15 15 45
#> AMERICAN INDIAN OR ALASKA NATIVE 0 0 1 (6.7%) 1 (2.2%)
#> ASIAN 15 (100%) 13 (86.7%) 8 (53.3%) 36 (80.0%)
#> BLACK OR AFRICAN AMERICAN 0 0 4 (26.7%) 4 (8.9%)
#> WHITE 0 2 (13.3%) 2 (13.3%) 4 (8.9%)4. Testing the codes for plot generation
The run function when calling a Graphics Template ID
returns a gTree object which will be used in the downstream
workflow for output generation. There are two alternative approaches to
rendering the plot: (1) having draw = TRUE in the
run function to enable the generated plot to be
automatically created and viewed via the Plots tab, and (2)
calling the function grid.draw from the package
grid which can be utilized to render the plot for viewing
and testing purpose. See example below:
proc_data <- log_filter(syn_data, PARAMCD == "OS", "adtte")
# method 1
run(kmg01, proc_data, dataset = "adtte", draw = TRUE)
# method 2
res <- run(kmg01, proc_data, dataset = "adtte")
grid::grid.newpage()
grid::grid.draw(res)General Control Arguments
1. lbl_overall: Column of Total
The generic argument lbl_overall controls whether the
column of total will be produced or not. lbl_overall = NULL
suppresses the total, lbl_overall = "All Patients" produces
the total.
2. Column counts: N=xxx
Column counts are displayed by default. There is no generic argument
controlling whether the count of unique number of subjects (N=xxx) will
be displayed in the column header or not. Users are allowed to customize
the display of N=xxx by forcing
display_columncounts = FALSE to wipe column counts away
during the postprocessing (with precautions and it is not
recommended).
tbl <- run(dmt01, syn_data) # table with column counts
tbl@col_info@display_columncounts <- FALSE
tbl # no column counts now
#> A: Drug X B: Placebo C: Combination All Patients
#> (N=15) (N=15) (N=15) (N=45)
#> ————————————————————————————————————————————————————————————————————————————————————————————
#> Age (yr)
#> n 15 15 15 45
#> Mean (SD) 31.3 (5.3) 35.1 (9.0) 36.6 (6.4) 34.3 (7.3)
#> Median 31.0 35.0 35.0 34.0
#> Min - Max 24 - 40 24 - 57 24 - 49 24 - 57
#> Age Group
#> n 15 15 15 45
#> <65 15 (100%) 15 (100%) 15 (100%) 45 (100%)
#> Sex
#> n 15 15 15 45
#> Male 3 (20.0%) 7 (46.7%) 5 (33.3%) 15 (33.3%)
#> Female 12 (80.0%) 8 (53.3%) 10 (66.7%) 30 (66.7%)
#> Ethnicity
#> n 15 15 15 45
#> HISPANIC OR LATINO 2 (13.3%) 0 0 2 (4.4%)
#> NOT HISPANIC OR LATINO 13 (86.7%) 15 (100%) 13 (86.7%) 41 (91.1%)
#> NOT REPORTED 0 0 2 (13.3%) 2 (4.4%)
#> RACE
#> n 15 15 15 45
#> AMERICAN INDIAN OR ALASKA NATIVE 0 2 (13.3%) 1 (6.7%) 3 (6.7%)
#> ASIAN 8 (53.3%) 10 (66.7%) 8 (53.3%) 26 (57.8%)
#> BLACK OR AFRICAN AMERICAN 4 (26.7%) 1 (6.7%) 4 (26.7%) 9 (20.0%)
#> WHITE 3 (20.0%) 2 (13.3%) 2 (13.3%) 7 (15.6%)TABLES
Safety Summary (AET01)
1. Safety Summary
The aet01 template produces the
standard safety summary.
run(aet01, syn_data, arm_var = "ARM")
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one AE 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of AEs 58 59 99
#> Total number of deaths 2 (13.3%) 4 (26.7%) 3 (20.0%)
#> Total number of patients withdrawn from study due to an AE 0 0 1 (6.7%)
#> Total number of patients with at least one
#> AE with fatal outcome 8 (53.3%) 8 (53.3%) 10 (66.7%)
#> Serious AE 12 (80.0%) 12 (80.0%) 11 (73.3%)
#> Serious AE leading to withdrawal from treatment 0 0 2 (13.3%)
#> Serious AE leading to dose modification/interruption 4 (26.7%) 3 (20.0%) 4 (26.7%)
#> Related Serious AE 8 (53.3%) 8 (53.3%) 10 (66.7%)
#> AE leading to withdrawal from treatment 2 (13.3%) 3 (20.0%) 3 (20.0%)
#> AE leading to dose modification/interruption 6 (40.0%) 9 (60.0%) 11 (73.3%)
#> Related AE 11 (73.3%) 10 (66.7%) 13 (86.7%)
#> Related AE leading to withdrawal from treatment 0 3 (20.0%) 0
#> Related AE leading to dose modification/interruption 1 (6.7%) 4 (26.7%) 9 (60.0%)
#> Severe AE (at greatest intensity) 11 (73.3%) 10 (66.7%) 12 (80.0%)2. Safety Summary with Modified Rows
Analyses under “Total number of patients with at least one” can be
removed, added, or modified by editing the parameter
anl_vars. An analysis here is an abbreviated name of the
analysis of interest, and supported by a variable in ADAE
derived under the condition of interest. The defined analyses currently
include "FATAL", "SER", "SERWD",
"SERDSM", "RELSER", "WD",
"DSM", "REL", "RELWD",
"RELDSM", and "SEV". When modification is
made, analyses must all be listed in the argument anl_vars.
The example below shows adding the customized analysis
"RELCTC35".
proc_data <- syn_data
proc_data$adae <- proc_data$adae %>%
filter(.data$ANL01FL == "Y") %>%
mutate(
FATAL = with_label(.data$AESDTH == "Y", "AE with fatal outcome"),
SER = with_label(.data$AESER == "Y", "Serious AE"),
SEV = with_label(.data$ASEV == "SEVERE", "Severe AE (at greatest intensity)"),
REL = with_label(.data$AREL == "Y", "Related AE"),
WD = with_label(.data$AEACN == "DRUG WITHDRAWN", "AE leading to withdrawal from treatment"),
DSM = with_label(
.data$AEACN %in% c("DRUG INTERRUPTED", "DOSE INCREASED", "DOSE REDUCED"),
"AE leading to dose modification/interruption"
),
SERWD = with_label(.data$SER & .data$WD, "Serious AE leading to withdrawal from treatment"),
SERDSM = with_label(.data$SER & .data$DSM, "Serious AE leading to dose modification/interruption"),
RELSER = with_label(.data$SER & .data$REL, "Related Serious AE"),
RELWD = with_label(.data$REL & .data$WD, "Related AE leading to withdrawal from treatment"),
RELDSM = with_label(.data$REL & .data$DSM, "Related AE leading to dose modification/interruption"),
CTC35 = with_label(.data$ATOXGR %in% c("3", "4", "5"), "Grade 3-5 AE"),
CTC45 = with_label(.data$ATOXGR %in% c("4", "5"), "Grade 4/5 AE"),
RELCTC35 = with_label(.data$ATOXGR %in% c("3", "4", "5") & .data$AEREL == "Y", "Related Grade 3-5")
)
proc_data$adsl <- proc_data$adsl %>%
mutate(DCSREAS = reformat(.data$DCSREAS, missing_rule))
run(aet01, proc_data, anl_vars = list(safety_var = c("FATAL", "SER", "RELSER", "RELCTC35")), auto_pre = FALSE)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one AE 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of AEs 58 59 99
#> Total number of deaths 2 (13.3%) 4 (26.7%) 3 (20.0%)
#> Total number of patients withdrawn from study due to an AE 0 0 1 (6.7%)
#> Total number of patients with at least one
#> AE with fatal outcome 8 (53.3%) 8 (53.3%) 10 (66.7%)
#> Serious AE 12 (80.0%) 12 (80.0%) 11 (73.3%)
#> Related Serious AE 8 (53.3%) 8 (53.3%) 10 (66.7%)
#> Related Grade 3-5 11 (73.3%) 10 (66.7%) 12 (80.0%)
Safety Summary (Adverse Events of Special Interest)
(AET01_AESI)
1. Safety Summary (Adverse Events of Special Interest)
The aet01_aesi template produces the
standard safety summary for adverse events of special interest.
run(aet01_aesi, syn_data)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one AE 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of AEs 58 59 99
#> Total number of patients with at least one AE by worst grade
#> Grade 1 0 1 (6.7%) 1 (6.7%)
#> Grade 2 1 (6.7%) 1 (6.7%) 1 (6.7%)
#> Grade 3 1 (6.7%) 2 (13.3%) 1 (6.7%)
#> Grade 4 3 (20.0%) 2 (13.3%) 2 (13.3%)
#> Grade 5 (fatal outcome) 8 (53.3%) 8 (53.3%) 10 (66.7%)
#> Total number of patients with study drug withdrawn due to AE 2 (13.3%) 3 (20.0%) 3 (20.0%)
#> Total number of patients with dose modified/interrupted due to AE 6 (40.0%) 9 (60.0%) 11 (73.3%)
#> Total number of patients with treatment received for AE 10 (66.7%) 10 (66.7%) 14 (93.3%)
#> Total number of patients with all non-fatal AEs resolved 9 (60.0%) 10 (66.7%) 12 (80.0%)
#> Total number of patients with at least one unresolved or ongoing non-fatal AE 10 (66.7%) 9 (60.0%) 14 (93.3%)
#> Total number of patients with at least one serious AE 12 (80.0%) 12 (80.0%) 11 (73.3%)
#> Total number of patients with at least one related AE 11 (73.3%) 10 (66.7%) 13 (86.7%)2. Safety Summary (Adverse Events of Special Interest) (optional lines)
Additional analyses can be added with the argument
aesi_vars, please type ?aet01_aesi in console
to find out the list of all pre-defined optional analyses in the
HELP.
run(aet01_aesi, syn_data, aesi_vars = c("RESLWD", "RELSER"))
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one AE 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of AEs 58 59 99
#> Total number of patients with at least one AE by worst grade
#> Grade 1 0 1 (6.7%) 1 (6.7%)
#> Grade 2 1 (6.7%) 1 (6.7%) 1 (6.7%)
#> Grade 3 1 (6.7%) 2 (13.3%) 1 (6.7%)
#> Grade 4 3 (20.0%) 2 (13.3%) 2 (13.3%)
#> Grade 5 (fatal outcome) 8 (53.3%) 8 (53.3%) 10 (66.7%)
#> Total number of patients with study drug withdrawn due to AE 2 (13.3%) 3 (20.0%) 3 (20.0%)
#> Total number of patients with dose modified/interrupted due to AE 6 (40.0%) 9 (60.0%) 11 (73.3%)
#> Total number of patients with treatment received for AE 10 (66.7%) 10 (66.7%) 14 (93.3%)
#> Total number of patients with all non-fatal AEs resolved 9 (60.0%) 10 (66.7%) 12 (80.0%)
#> Total number of patients with at least one unresolved or ongoing non-fatal AE 10 (66.7%) 9 (60.0%) 14 (93.3%)
#> Total number of patients with at least one serious AE 12 (80.0%) 12 (80.0%) 11 (73.3%)
#> Total number of patients with at least one related AE 11 (73.3%) 10 (66.7%) 13 (86.7%)
#> No. of patients with serious, related AE 8 (53.3%) 8 (53.3%) 10 (66.7%)
Adverse Events (AET02)
1. Adverse Events
- The template
aet02produces the standard adverse event summary by MedDRA system organ class and preferred term. - The template does not include the column of total as default. The
‘All Patients’ column can be added with the argument
lbl_overall = "All Patients". - Missing values in
"AEBODSYS", and"AEDECOD"are labeled asNo Coding Available.
run(aet02, syn_data)
#> MedDRA System Organ Class A: Drug X B: Placebo C: Combination
#> MedDRA Preferred Term (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one adverse event 13 (86.7%) 14 (93.3%) 15 (100%)
#> Overall total number of events 58 59 99
#> cl B.2
#> Total number of patients with at least one adverse event 11 (73.3%) 8 (53.3%) 10 (66.7%)
#> Total number of events 18 15 20
#> dcd B.2.2.3.1 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> dcd B.2.1.2.1 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> cl D.1
#> Total number of patients with at least one adverse event 9 (60.0%) 5 (33.3%) 11 (73.3%)
#> Total number of events 13 9 19
#> dcd D.1.1.1.1 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.4.2 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> cl A.1
#> Total number of patients with at least one adverse event 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> Total number of events 8 11 16
#> dcd A.1.1.1.2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> dcd A.1.1.1.1 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> cl B.1
#> Total number of patients with at least one adverse event 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> Total number of events 6 6 12
#> dcd B.1.1.1.1 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> cl C.2
#> Total number of patients with at least one adverse event 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Total number of events 6 4 12
#> dcd C.2.1.2.1 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> cl D.2
#> Total number of patients with at least one adverse event 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> Total number of events 3 5 10
#> dcd D.2.1.5.3 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> cl C.1
#> Total number of patients with at least one adverse event 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Total number of events 4 9 10
#> dcd C.1.1.1.3 4 (26.7%) 4 (26.7%) 5 (33.3%)2. Adverse Events (with High-level Term)
The syntax below displays adverse events by MedDRA system organ class, high-level term and preferred term.
run(aet02, syn_data, row_split_var = c("AEBODSYS", "AEHLT"))
#> MedDRA System Organ Class
#> High Level Term A: Drug X B: Placebo C: Combination
#> MedDRA Preferred Term (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one adverse event 13 (86.7%) 14 (93.3%) 15 (100%)
#> Overall total number of events 58 59 99
#> cl B.2
#> Total number of patients with at least one adverse event 11 (73.3%) 8 (53.3%) 10 (66.7%)
#> Total number of events 18 15 20
#> hlt B.2.2.3
#> Total number of patients with at least one adverse event 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> Total number of events 9 7 13
#> dcd B.2.2.3.1 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> hlt B.2.1.2
#> Total number of patients with at least one adverse event 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> Total number of events 9 8 7
#> dcd B.2.1.2.1 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> cl D.1
#> Total number of patients with at least one adverse event 9 (60.0%) 5 (33.3%) 11 (73.3%)
#> Total number of events 13 9 19
#> hlt D.1.1.1
#> Total number of patients with at least one adverse event 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> Total number of events 5 7 11
#> dcd D.1.1.1.1 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> hlt D.1.1.4
#> Total number of patients with at least one adverse event 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> Total number of events 8 2 8
#> dcd D.1.1.4.2 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> cl A.1
#> Total number of patients with at least one adverse event 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> Total number of events 8 11 16
#> hlt A.1.1.1
#> Total number of patients with at least one adverse event 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> Total number of events 8 11 16
#> dcd A.1.1.1.2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> dcd A.1.1.1.1 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> cl B.1
#> Total number of patients with at least one adverse event 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> Total number of events 6 6 12
#> hlt B.1.1.1
#> Total number of patients with at least one adverse event 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> Total number of events 6 6 12
#> dcd B.1.1.1.1 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> cl C.2
#> Total number of patients with at least one adverse event 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Total number of events 6 4 12
#> hlt C.2.1.2
#> Total number of patients with at least one adverse event 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Total number of events 6 4 12
#> dcd C.2.1.2.1 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> cl D.2
#> Total number of patients with at least one adverse event 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> Total number of events 3 5 10
#> hlt D.2.1.5
#> Total number of patients with at least one adverse event 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> Total number of events 3 5 10
#> dcd D.2.1.5.3 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> cl C.1
#> Total number of patients with at least one adverse event 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Total number of events 4 9 10
#> hlt C.1.1.1
#> Total number of patients with at least one adverse event 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Total number of events 4 9 10
#> dcd C.1.1.1.3 4 (26.7%) 4 (26.7%) 5 (33.3%)3. Adverse Events (Preferred Terms only)
The syntax below displays adverse events by preferred term only.
run(aet02, syn_data, row_split_var = NULL)
#> A: Drug X B: Placebo C: Combination
#> MedDRA Preferred Term (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one adverse event 13 (86.7%) 14 (93.3%) 15 (100%)
#> Overall total number of events 58 59 99
#> dcd B.2.2.3.1 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> dcd B.1.1.1.1 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> dcd C.2.1.2.1 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> dcd A.1.1.1.2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> dcd B.2.1.2.1 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> dcd D.1.1.1.1 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.4.2 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> dcd D.2.1.5.3 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> dcd C.1.1.1.3 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> dcd A.1.1.1.1 3 (20.0%) 1 (6.7%) 6 (40.0%)
Adverse Events by Greatest
Intensity(AET03)
1. Adverse Events by Greatest Intensity
This aet03 template produces the
standard adverse event by greatest intensity summary
run(aet03, syn_data)
#> MedDRA System Organ Class A: Drug X B: Placebo C: Combination
#> MedDRA Preferred Term (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————
#> - Any Intensity - 13 (86.7%) 14 (93.3%) 15 (100%)
#> MILD 0 1 (6.7%) 1 (6.7%)
#> MODERATE 2 (13.3%) 3 (20.0%) 2 (13.3%)
#> SEVERE 11 (73.3%) 10 (66.7%) 12 (80.0%)
#> cl B.2
#> - Any Intensity - 11 (73.3%) 8 (53.3%) 10 (66.7%)
#> MILD 6 (40.0%) 2 (13.3%) 5 (33.3%)
#> MODERATE 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> dcd B.2.2.3.1
#> - Any Intensity - 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> MILD 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> dcd B.2.1.2.1
#> - Any Intensity - 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> MODERATE 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> cl D.1
#> - Any Intensity - 9 (60.0%) 5 (33.3%) 11 (73.3%)
#> MODERATE 5 (33.3%) 1 (6.7%) 4 (26.7%)
#> SEVERE 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.1.1
#> - Any Intensity - 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> SEVERE 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.4.2
#> - Any Intensity - 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> MODERATE 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> cl A.1
#> - Any Intensity - 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> MILD 2 (13.3%) 0 4 (26.7%)
#> MODERATE 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> dcd A.1.1.1.2
#> - Any Intensity - 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> MODERATE 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> dcd A.1.1.1.1
#> - Any Intensity - 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> MILD 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> cl B.1
#> - Any Intensity - 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> SEVERE 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> dcd B.1.1.1.1
#> - Any Intensity - 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> SEVERE 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> cl C.2
#> - Any Intensity - 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> MODERATE 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> dcd C.2.1.2.1
#> - Any Intensity - 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> MODERATE 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> cl D.2
#> - Any Intensity - 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> MILD 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> dcd D.2.1.5.3
#> - Any Intensity - 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> MILD 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> cl C.1
#> - Any Intensity - 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> SEVERE 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> dcd C.1.1.1.3
#> - Any Intensity - 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> SEVERE 4 (26.7%) 4 (26.7%) 5 (33.3%)
Adverse Events by Highest NCI CTCAE Grade
(AET04)
1. Adverse Events by Highest NCI CTCAE
Grade
- The
aet04template produces the standard adverse event by highestNCI CTCAEgrade summary. - By default, this template includes the grouped grades of ‘Grade 1-2’ and ‘Grade 3-4’.
- By default this template removes the rows with 0 count.
- If a treatment group does not have any adverse event, the treatment
group is automatically displayed providing that it is defined in
ADSL.
run(aet04, syn_data)
#> MedDRA System Organ Class
#> MedDRA Preferred Term A: Drug X B: Placebo C: Combination
#> Grade (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————
#> - Any adverse events -
#> - Any Grade - 13 (86.7%) 14 (93.3%) 15 (100%)
#> Grade 1-2 1 (6.7%) 2 (13.3%) 2 (13.3%)
#> 1 0 1 (6.7%) 1 (6.7%)
#> 2 1 (6.7%) 1 (6.7%) 1 (6.7%)
#> Grade 3-4 4 (26.7%) 4 (26.7%) 3 (20.0%)
#> 3 1 (6.7%) 2 (13.3%) 1 (6.7%)
#> 4 3 (20.0%) 2 (13.3%) 2 (13.3%)
#> Grade 5 8 (53.3%) 8 (53.3%) 10 (66.7%)
#> cl B.2
#> - Overall -
#> - Any Grade - 11 (73.3%) 8 (53.3%) 10 (66.7%)
#> Grade 1-2 6 (40.0%) 2 (13.3%) 5 (33.3%)
#> 1 6 (40.0%) 2 (13.3%) 5 (33.3%)
#> Grade 3-4 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 3 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> dcd B.2.2.3.1
#> - Any Grade - 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> Grade 1-2 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> 1 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> dcd B.2.1.2.1
#> - Any Grade - 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> Grade 3-4 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 3 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> cl D.1
#> - Overall -
#> - Any Grade - 9 (60.0%) 5 (33.3%) 11 (73.3%)
#> Grade 3-4 5 (33.3%) 1 (6.7%) 4 (26.7%)
#> 3 5 (33.3%) 1 (6.7%) 4 (26.7%)
#> Grade 5 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.1.1
#> - Any Grade - 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> Grade 5 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.4.2
#> - Any Grade - 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> Grade 3-4 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> 3 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> cl A.1
#> - Overall -
#> - Any Grade - 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> Grade 1-2 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> 1 2 (13.3%) 0 4 (26.7%)
#> 2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> dcd A.1.1.1.2
#> - Any Grade - 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> Grade 1-2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> 2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> dcd A.1.1.1.1
#> - Any Grade - 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> Grade 1-2 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> 1 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> cl B.1
#> - Overall -
#> - Any Grade - 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> Grade 5 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> dcd B.1.1.1.1
#> - Any Grade - 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> Grade 5 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> cl C.2
#> - Overall -
#> - Any Grade - 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 1-2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> 2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> dcd C.2.1.2.1
#> - Any Grade - 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 1-2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> 2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> cl D.2
#> - Overall -
#> - Any Grade - 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> Grade 1-2 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 1 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> dcd D.2.1.5.3
#> - Any Grade - 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> Grade 1-2 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 1 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> cl C.1
#> - Overall -
#> - Any Grade - 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Grade 3-4 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> 4 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> dcd C.1.1.1.3
#> - Any Grade - 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Grade 3-4 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> 4 4 (26.7%) 4 (26.7%) 5 (33.3%)
2. Adverse Events by Highest NCI CTCAE Grade
(Fill in of Grades)
If, for some preferred terms, not all grades occur but all grades
should be displayed, this can be achieved by specifying the argument
prune_0 = FALSE.
run(aet04, syn_data, prune_0 = FALSE)
#> MedDRA System Organ Class
#> MedDRA Preferred Term A: Drug X B: Placebo C: Combination
#> Grade (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————
#> - Any adverse events -
#> - Any Grade - 13 (86.7%) 14 (93.3%) 15 (100%)
#> Grade 1-2 1 (6.7%) 2 (13.3%) 2 (13.3%)
#> 1 0 1 (6.7%) 1 (6.7%)
#> 2 1 (6.7%) 1 (6.7%) 1 (6.7%)
#> Grade 3-4 4 (26.7%) 4 (26.7%) 3 (20.0%)
#> 3 1 (6.7%) 2 (13.3%) 1 (6.7%)
#> 4 3 (20.0%) 2 (13.3%) 2 (13.3%)
#> Grade 5 8 (53.3%) 8 (53.3%) 10 (66.7%)
#> cl B.2
#> - Overall -
#> - Any Grade - 11 (73.3%) 8 (53.3%) 10 (66.7%)
#> Grade 1-2 6 (40.0%) 2 (13.3%) 5 (33.3%)
#> 1 6 (40.0%) 2 (13.3%) 5 (33.3%)
#> 2 0 0 0
#> Grade 3-4 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 3 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 4 0 0 0
#> Grade 5 0 0 0
#> dcd B.2.2.3.1
#> - Any Grade - 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> Grade 1-2 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> 1 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> 2 0 0 0
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 0 0 0
#> dcd B.2.1.2.1
#> - Any Grade - 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-4 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 3 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 4 0 0 0
#> Grade 5 0 0 0
#> cl D.1
#> - Overall -
#> - Any Grade - 9 (60.0%) 5 (33.3%) 11 (73.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-4 5 (33.3%) 1 (6.7%) 4 (26.7%)
#> 3 5 (33.3%) 1 (6.7%) 4 (26.7%)
#> 4 0 0 0
#> Grade 5 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.1.1
#> - Any Grade - 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.4.2
#> - Any Grade - 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-4 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> 3 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> 4 0 0 0
#> Grade 5 0 0 0
#> cl A.1
#> - Overall -
#> - Any Grade - 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> Grade 1-2 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> 1 2 (13.3%) 0 4 (26.7%)
#> 2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 0 0 0
#> dcd A.1.1.1.2
#> - Any Grade - 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> Grade 1-2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> 1 0 0 0
#> 2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 0 0 0
#> dcd A.1.1.1.1
#> - Any Grade - 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> Grade 1-2 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> 1 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> 2 0 0 0
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 0 0 0
#> cl B.1
#> - Overall -
#> - Any Grade - 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> dcd B.1.1.1.1
#> - Any Grade - 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> cl C.2
#> - Overall -
#> - Any Grade - 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 1-2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> 1 0 0 0
#> 2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 0 0 0
#> dcd C.2.1.2.1
#> - Any Grade - 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 1-2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> 1 0 0 0
#> 2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 0 0 0
#> cl D.2
#> - Overall -
#> - Any Grade - 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> Grade 1-2 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 1 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 2 0 0 0
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 0 0 0
#> dcd D.2.1.5.3
#> - Any Grade - 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> Grade 1-2 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 1 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 2 0 0 0
#> Grade 3-4 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Grade 5 0 0 0
#> cl C.1
#> - Overall -
#> - Any Grade - 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-4 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> 3 0 0 0
#> 4 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Grade 5 0 0 0
#> dcd C.1.1.1.3
#> - Any Grade - 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-4 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> 3 0 0 0
#> 4 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Grade 5 0 0 0
3. Adverse Events by Highest NCI CTCAE Grade
with modified grouping of grade
Collapsing grade 3-4 with grade 5, can be achieved by modifying the
definition of grade groups in the argument
grade_groups.
grade_groups <- list(
"Grade 1-2" = c("1", "2"),
"Grade 3-5" = c("3", "4", "5")
)
run(aet04, syn_data, grade_groups = grade_groups, prune_0 = FALSE)
#> MedDRA System Organ Class
#> MedDRA Preferred Term A: Drug X B: Placebo C: Combination
#> Grade (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————
#> - Any adverse events -
#> - Any Grade - 13 (86.7%) 14 (93.3%) 15 (100%)
#> Grade 1-2 1 (6.7%) 2 (13.3%) 2 (13.3%)
#> 1 0 1 (6.7%) 1 (6.7%)
#> 2 1 (6.7%) 1 (6.7%) 1 (6.7%)
#> Grade 3-5 12 (80.0%) 12 (80.0%) 13 (86.7%)
#> 3 1 (6.7%) 2 (13.3%) 1 (6.7%)
#> 4 3 (20.0%) 2 (13.3%) 2 (13.3%)
#> 5 8 (53.3%) 8 (53.3%) 10 (66.7%)
#> cl B.2
#> - Overall -
#> - Any Grade - 11 (73.3%) 8 (53.3%) 10 (66.7%)
#> Grade 1-2 6 (40.0%) 2 (13.3%) 5 (33.3%)
#> 1 6 (40.0%) 2 (13.3%) 5 (33.3%)
#> 2 0 0 0
#> Grade 3-5 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 3 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 4 0 0 0
#> 5 0 0 0
#> dcd B.2.2.3.1
#> - Any Grade - 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> Grade 1-2 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> 1 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> 2 0 0 0
#> Grade 3-5 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> 5 0 0 0
#> dcd B.2.1.2.1
#> - Any Grade - 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-5 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 3 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> 4 0 0 0
#> 5 0 0 0
#> cl D.1
#> - Overall -
#> - Any Grade - 9 (60.0%) 5 (33.3%) 11 (73.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-5 9 (60.0%) 5 (33.3%) 11 (73.3%)
#> 3 5 (33.3%) 1 (6.7%) 4 (26.7%)
#> 4 0 0 0
#> 5 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.1.1
#> - Any Grade - 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-5 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> 3 0 0 0
#> 4 0 0 0
#> 5 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.4.2
#> - Any Grade - 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-5 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> 3 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> 4 0 0 0
#> 5 0 0 0
#> cl A.1
#> - Overall -
#> - Any Grade - 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> Grade 1-2 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> 1 2 (13.3%) 0 4 (26.7%)
#> 2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> Grade 3-5 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> 5 0 0 0
#> dcd A.1.1.1.2
#> - Any Grade - 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> Grade 1-2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> 1 0 0 0
#> 2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> Grade 3-5 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> 5 0 0 0
#> dcd A.1.1.1.1
#> - Any Grade - 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> Grade 1-2 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> 1 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> 2 0 0 0
#> Grade 3-5 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> 5 0 0 0
#> cl B.1
#> - Overall -
#> - Any Grade - 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-5 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> 3 0 0 0
#> 4 0 0 0
#> 5 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> dcd B.1.1.1.1
#> - Any Grade - 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-5 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> 3 0 0 0
#> 4 0 0 0
#> 5 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> cl C.2
#> - Overall -
#> - Any Grade - 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 1-2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> 1 0 0 0
#> 2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 3-5 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> 5 0 0 0
#> dcd C.2.1.2.1
#> - Any Grade - 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 1-2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> 1 0 0 0
#> 2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> Grade 3-5 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> 5 0 0 0
#> cl D.2
#> - Overall -
#> - Any Grade - 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> Grade 1-2 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 1 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 2 0 0 0
#> Grade 3-5 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> 5 0 0 0
#> dcd D.2.1.5.3
#> - Any Grade - 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> Grade 1-2 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 1 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> 2 0 0 0
#> Grade 3-5 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> 5 0 0 0
#> cl C.1
#> - Overall -
#> - Any Grade - 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-5 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> 3 0 0 0
#> 4 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> 5 0 0 0
#> dcd C.1.1.1.3
#> - Any Grade - 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> Grade 1-2 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> Grade 3-5 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> 3 0 0 0
#> 4 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> 5 0 0 0
Adverse Event Rate Adjusted for Patient-Years at Risk -
First Occurrence (AET05)
1. Adverse Event Rate Adjusted for Patient-Years at Risk - First Occurrence
- The
aet05template produces the standard adverse event rate adjusted for patient-years at risk summary considering first occurrence only. - By default, all
adsaftteparameter codes containing the string"TTE"are included in the output. Users are expected to filter the parameter(s) of interest from input safety time-to-event dataset in pre-processing if needed. - In the input safety time-to-event dataset, in the censoring variable
CNSR,0indicates the occurrence of an event of interest and1denotes censoring.
proc_data <- log_filter(syn_data, PARAMCD == "AETTE1", "adsaftte")
run(aet05, proc_data)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————————————————
#> Time to first occurrence of any adverse event
#> Total patient-years at risk 31.0 9.0 22.0
#> Number of adverse events observed 5 13 8
#> AE rate per 100 patient-years 16.13 143.75 36.30
#> 95% CI (1.99, 30.27) (65.61, 221.89) (11.15, 61.45)2. Adverse Event Rate Adjusted for Patient-Years at Risk - First Occurrence (setting type of confidence interval)
- The type of the confidence interval for rate can be specified by the
argument
conf_type. Options includenormal(default),normal_logandexact. - The confidence interval can be adjusted by the argument
conf_level.
run(aet05, syn_data, conf_level = 0.90, conf_type = "exact")
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Time to first occurrence of a grade 3-5 adverse event
#> Total patient-years at risk 10.3 6.3 8.3
#> Number of adverse events observed 12 14 13
#> AE rate per 100 patient-years 116.36 223.74 156.98
#> 90% CI (67.14, 188.53) (135.27, 349.78) (92.86, 249.59)
#> Time to first occurrence of any adverse event
#> Total patient-years at risk 31.0 9.0 22.0
#> Number of adverse events observed 5 13 8
#> AE rate per 100 patient-years 16.13 143.75 36.30
#> 90% CI (6.36, 33.91) (85.03, 228.55) (18.06, 65.50)
#> Time to first occurrence of any serious adverse event
#> Total patient-years at risk 32.9 7.6 9.4
#> Number of adverse events observed 4 14 13
#> AE rate per 100 patient-years 12.15 183.83 137.79
#> 90% CI (4.15, 27.80) (111.14, 287.38) (81.50, 219.06)
Adverse Event Rate Adjusted for Patient-Years at Risk - All
Occurrences (AET05_ALL)
1. Adverse Event Rate Adjusted for Patient-Years at Risk - All Occurrences
- The
aet05_alltemplate produces the standard adverse event rate adjusted for patient-years at risk summary considering all occurrences. - By default, all
adsaftteparameter codes containing the string"TOT"and the parameter code"AEREPTTE"are required."TOT"parameters store the number of occurrences of adverse event of interests. Parameter code"AEREPTTE"stores the time to end of adverse event reporting period in years that contribute to the summary of “total patient-years at risk” in the output. Users are expected to filter parameters of interest from input analysis dataset in pre-processing, if needed. - In the input safety time-to-event dataset, in the censoring variable
CNSR,0indicates the occurrence of an event of interest and1denotes censoring.
proc_data <- log_filter(syn_data, PARAMCD == "AETOT1" | PARAMCD == "AEREPTTE", "adsaftte")
run(aet05_all, proc_data)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————————————————
#> Number of occurrences of any adverse event
#> Total patient-years at risk 44.4 44.2 44.4
#> Number of adverse events observed 29 49 56
#> AE rate per 100 patient-years 65.32 110.76 126.15
#> 95% CI (41.54, 89.09) (79.75, 141.77) (93.11, 159.19)2. Adverse Event Rate Adjusted for Patient-Years at Risk - All Occurrences (setting type of confidence interval)
- The type of the confidence interval for rate can be specified by the
argument
conf_type. Options includenormal(default),normal_log,exact, andbyar. - The confidence interval can be adjusted by the argument
conf_level.
run(aet05_all, syn_data, conf_level = 0.90, conf_type = "exact")
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ——————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Number of occurrences of a grade 3-5 adverse event
#> Total patient-years at risk 44.4 44.2 44.4
#> Number of adverse events observed 65 54 95
#> AE rate per 100 patient-years 146.40 122.06 214.00
#> 90% CI (117.86, 179.97) (96.08, 153.12) (179.22, 253.80)
#> Number of occurrences of any adverse event
#> Total patient-years at risk 44.4 44.2 44.4
#> Number of adverse events observed 29 49 56
#> AE rate per 100 patient-years 65.32 110.76 126.15
#> 90% CI (46.73, 89.06) (86.08, 140.53) (99.76, 157.60)
#> Number of occurrences of any serious adverse event
#> Total patient-years at risk 44.4 44.2 44.4
#> Number of adverse events observed 9 36 60
#> AE rate per 100 patient-years 20.27 81.37 135.16
#> 90% CI (10.57, 35.37) (60.42, 107.46) (107.80, 167.58)
Most Common (>=5%) Adverse Events
(AET10)
1. Most Common (>=5%) Adverse Events
- The
aet10template produces the standard most common adverse events occurring with relative frequency >=5% output.
run(aet10, syn_data)
#> A: Drug X B: Placebo C: Combination
#> MedDRA Preferred Term (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————
#> dcd B.2.2.3.1 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> dcd B.1.1.1.1 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> dcd C.2.1.2.1 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> dcd A.1.1.1.2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> dcd B.2.1.2.1 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> dcd D.1.1.1.1 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.4.2 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> dcd D.2.1.5.3 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> dcd C.1.1.1.3 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> dcd A.1.1.1.1 3 (20.0%) 1 (6.7%) 6 (40.0%)2. Most Common (>=8%) Adverse Events (setting threshold)
To modify the threshold for displaying preferred terms, this can be
achieved by providing the threshold to the argument
atleast.
run(aet10, syn_data, atleast = 0.08)
#> A: Drug X B: Placebo C: Combination
#> MedDRA Preferred Term (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————
#> dcd B.2.2.3.1 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> dcd B.1.1.1.1 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> dcd C.2.1.2.1 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> dcd A.1.1.1.2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> dcd B.2.1.2.1 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> dcd D.1.1.1.1 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> dcd D.1.1.4.2 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> dcd D.2.1.5.3 2 (13.3%) 5 (33.3%) 7 (46.7%)
#> dcd C.1.1.1.3 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> dcd A.1.1.1.1 3 (20.0%) 1 (6.7%) 6 (40.0%)
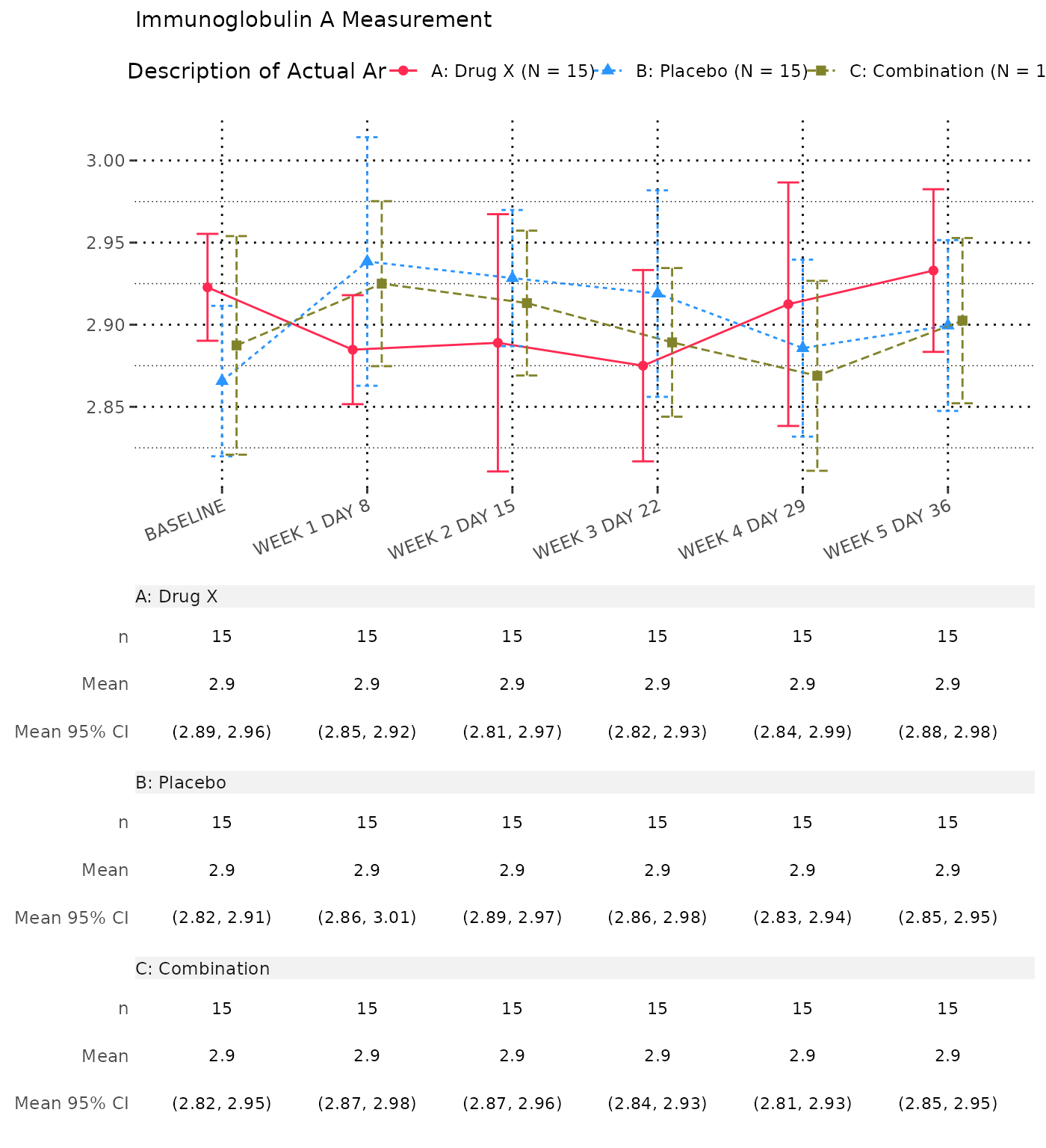
Absolute Value and Change from Baseline by Visit
(CFBT01)
1. Absolute Value and Change from Baseline by Visit
- By default, the
cfbt01template displays analysis value (AVAL) and absolute change from baseline (CHG) for each visit. - The template does not include the column of total by default.
- Each parameter is presented on a separate page.
- The absolute change from baseline at baseline value is not displayed.
proc_data <- log_filter(
syn_data,
PARAMCD %in% c("DIABP", "SYSBP"), "advs"
)
run(cfbt01, proc_data, dataset = "advs")
#> A: Drug X B: Placebo C: Combination
#> Change from Change from Change from
#> Value at Visit Baseline Value at Visit Baseline Value at Visit Baseline
#> Analysis Visit (N=15) (N=15) (N=15) (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Diastolic Blood Pressure
#> SCREENING
#> n 15 0 15 0 15 0
#> Mean (SD) 94.385 (17.067) NE (NE) 106.381 (20.586) NE (NE) 106.468 (12.628) NE (NE)
#> Median 94.933 NE 111.133 NE 108.359 NE
#> Min - Max 55.71 - 122.00 NE - NE 60.21 - 131.91 NE - NE 83.29 - 127.17 NE - NE
#> BASELINE
#> n 15 15 15
#> Mean (SD) 96.133 (22.458) 108.111 (15.074) 103.149 (19.752)
#> Median 93.328 108.951 102.849
#> Min - Max 60.58 - 136.59 83.44 - 131.62 66.05 - 136.55
#> WEEK 1 DAY 8
#> n 15 15 15 15 15 15
#> Mean (SD) 98.977 (21.359) 2.844 (28.106) 104.110 (16.172) -4.001 (21.867) 100.826 (19.027) -2.323 (25.018)
#> Median 92.447 -4.066 107.703 3.227 103.058 -2.476
#> Min - Max 67.55 - 130.37 -32.82 - 47.68 70.91 - 132.89 -52.94 - 28.63 70.04 - 128.68 -55.15 - 41.81
#> WEEK 2 DAY 15
#> n 15 15 15 15 15 15
#> Mean (SD) 99.758 (14.477) 3.626 (21.189) 97.473 (17.296) -10.638 (20.831) 94.272 (16.961) -8.877 (27.229)
#> Median 101.498 1.731 99.501 -9.727 96.789 -10.155
#> Min - Max 71.98 - 122.81 -39.50 - 47.57 53.80 - 125.81 -55.15 - 25.26 63.45 - 117.47 -73.10 - 46.54
#> WEEK 3 DAY 22
#> n 15 15 15 15 15 15
#> Mean (SD) 99.101 (26.109) 2.968 (34.327) 91.984 (16.925) -16.127 (21.881) 94.586 (13.560) -8.563 (21.713)
#> Median 101.146 -0.271 91.244 -14.384 98.398 -16.075
#> Min - Max 47.68 - 162.22 -47.87 - 76.64 67.80 - 119.72 -53.06 - 22.52 73.50 - 115.43 -37.90 - 32.66
#> WEEK 4 DAY 29
#> n 15 15 15 15 15 15
#> Mean (SD) 103.400 (22.273) 7.267 (30.740) 96.467 (19.451) -11.644 (25.922) 108.338 (18.417) 5.189 (21.881)
#> Median 98.168 2.510 97.385 -16.793 107.555 7.966
#> Min - Max 63.09 - 148.25 -38.43 - 61.90 63.35 - 131.57 -57.11 - 48.13 68.78 - 132.52 -33.96 - 41.50
#> WEEK 5 DAY 36
#> n 15 15 15 15 15 15
#> Mean (SD) 93.222 (18.536) -2.911 (28.873) 97.890 (20.701) -10.221 (27.593) 95.317 (16.401) -7.832 (19.827)
#> Median 90.799 -3.385 99.049 -11.319 93.876 -4.665
#> Min - Max 63.55 - 139.11 -48.63 - 47.35 69.47 - 137.64 -54.38 - 37.85 71.91 - 138.54 -44.47 - 29.11
#> Systolic Blood Pressure
#> SCREENING
#> n 15 0 15 0 15 0
#> Mean (SD) 154.073 (33.511) NE (NE) 157.840 (34.393) NE (NE) 152.407 (22.311) NE (NE)
#> Median 156.169 NE 161.670 NE 149.556 NE
#> Min - Max 78.31 - 210.70 NE - NE 79.76 - 210.40 NE - NE 108.21 - 184.88 NE - NE
#> BASELINE
#> n 15 15 15
#> Mean (SD) 145.925 (28.231) 152.007 (28.664) 154.173 (26.317)
#> Median 142.705 157.698 155.282
#> Min - Max 85.21 - 195.68 98.90 - 194.62 86.65 - 192.68
#> WEEK 1 DAY 8
#> n 15 15 15 15 15 15
#> Mean (SD) 156.509 (21.097) 10.584 (34.598) 147.480 (33.473) -4.527 (48.895) 143.319 (30.759) -10.854 (34.553)
#> Median 160.711 5.802 155.030 2.758 145.548 -5.636
#> Min - Max 126.84 - 185.53 -53.28 - 91.52 85.22 - 189.88 -77.34 - 90.98 90.37 - 191.58 -65.71 - 49.04
#> WEEK 2 DAY 15
#> n 15 15 15 15 15 15
#> Mean (SD) 144.202 (33.676) -1.723 (27.067) 136.892 (30.178) -15.115 (37.794) 148.622 (27.088) -5.551 (44.670)
#> Median 144.253 5.325 142.679 -14.083 147.102 -11.512
#> Min - Max 62.56 - 203.66 -53.89 - 44.16 70.34 - 174.27 -83.07 - 62.39 108.82 - 200.23 -69.54 - 113.59
#> WEEK 3 DAY 22
#> n 15 15 15 15 15 15
#> Mean (SD) 154.887 (35.374) 8.962 (38.455) 149.761 (28.944) -2.247 (44.835) 150.460 (21.352) -3.712 (37.984)
#> Median 158.938 17.191 155.044 -1.796 156.505 -7.606
#> Min - Max 112.32 - 218.83 -47.28 - 96.18 84.42 - 192.92 -110.20 - 94.02 94.70 - 180.41 -74.91 - 72.74
#> WEEK 4 DAY 29
#> n 15 15 15 15 15 15
#> Mean (SD) 150.159 (32.249) 4.234 (32.965) 156.043 (22.863) 4.036 (42.494) 145.714 (22.980) -8.458 (33.175)
#> Median 145.506 3.754 149.094 -10.000 150.797 -14.432
#> Min - Max 69.37 - 210.43 -89.16 - 54.32 113.57 - 195.10 -71.44 - 77.75 106.91 - 188.09 -41.95 - 65.16
#> WEEK 5 DAY 36
#> n 15 15 15 15 15 15
#> Mean (SD) 155.964 (30.945) 10.039 (42.252) 156.387 (35.274) 4.380 (51.782) 143.592 (33.170) -10.581 (44.799)
#> Median 158.142 1.448 164.552 7.060 148.501 -2.385
#> Min - Max 110.61 - 212.47 -53.91 - 90.45 63.28 - 198.79 -131.34 - 86.84 92.18 - 191.05 -78.77 - 64.352. Absolute Value and Change from Baseline by Visit without Screening
The skip arguments controls which visit values should
not be displayed. For instance, to mask the changes from baseline during
the “SCREENING” and “BASELINE” visits.
run(cfbt01, proc_data, dataset = "advs", skip = list(CHG = c("SCREENING", "BASELINE")))
#> A: Drug X B: Placebo C: Combination
#> Change from Change from Change from
#> Value at Visit Baseline Value at Visit Baseline Value at Visit Baseline
#> Analysis Visit (N=15) (N=15) (N=15) (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Diastolic Blood Pressure
#> SCREENING
#> n 15 15 15
#> Mean (SD) 94.385 (17.067) 106.381 (20.586) 106.468 (12.628)
#> Median 94.933 111.133 108.359
#> Min - Max 55.71 - 122.00 60.21 - 131.91 83.29 - 127.17
#> BASELINE
#> n 15 15 15
#> Mean (SD) 96.133 (22.458) 108.111 (15.074) 103.149 (19.752)
#> Median 93.328 108.951 102.849
#> Min - Max 60.58 - 136.59 83.44 - 131.62 66.05 - 136.55
#> WEEK 1 DAY 8
#> n 15 15 15 15 15 15
#> Mean (SD) 98.977 (21.359) 2.844 (28.106) 104.110 (16.172) -4.001 (21.867) 100.826 (19.027) -2.323 (25.018)
#> Median 92.447 -4.066 107.703 3.227 103.058 -2.476
#> Min - Max 67.55 - 130.37 -32.82 - 47.68 70.91 - 132.89 -52.94 - 28.63 70.04 - 128.68 -55.15 - 41.81
#> WEEK 2 DAY 15
#> n 15 15 15 15 15 15
#> Mean (SD) 99.758 (14.477) 3.626 (21.189) 97.473 (17.296) -10.638 (20.831) 94.272 (16.961) -8.877 (27.229)
#> Median 101.498 1.731 99.501 -9.727 96.789 -10.155
#> Min - Max 71.98 - 122.81 -39.50 - 47.57 53.80 - 125.81 -55.15 - 25.26 63.45 - 117.47 -73.10 - 46.54
#> WEEK 3 DAY 22
#> n 15 15 15 15 15 15
#> Mean (SD) 99.101 (26.109) 2.968 (34.327) 91.984 (16.925) -16.127 (21.881) 94.586 (13.560) -8.563 (21.713)
#> Median 101.146 -0.271 91.244 -14.384 98.398 -16.075
#> Min - Max 47.68 - 162.22 -47.87 - 76.64 67.80 - 119.72 -53.06 - 22.52 73.50 - 115.43 -37.90 - 32.66
#> WEEK 4 DAY 29
#> n 15 15 15 15 15 15
#> Mean (SD) 103.400 (22.273) 7.267 (30.740) 96.467 (19.451) -11.644 (25.922) 108.338 (18.417) 5.189 (21.881)
#> Median 98.168 2.510 97.385 -16.793 107.555 7.966
#> Min - Max 63.09 - 148.25 -38.43 - 61.90 63.35 - 131.57 -57.11 - 48.13 68.78 - 132.52 -33.96 - 41.50
#> WEEK 5 DAY 36
#> n 15 15 15 15 15 15
#> Mean (SD) 93.222 (18.536) -2.911 (28.873) 97.890 (20.701) -10.221 (27.593) 95.317 (16.401) -7.832 (19.827)
#> Median 90.799 -3.385 99.049 -11.319 93.876 -4.665
#> Min - Max 63.55 - 139.11 -48.63 - 47.35 69.47 - 137.64 -54.38 - 37.85 71.91 - 138.54 -44.47 - 29.11
#> Systolic Blood Pressure
#> SCREENING
#> n 15 15 15
#> Mean (SD) 154.073 (33.511) 157.840 (34.393) 152.407 (22.311)
#> Median 156.169 161.670 149.556
#> Min - Max 78.31 - 210.70 79.76 - 210.40 108.21 - 184.88
#> BASELINE
#> n 15 15 15
#> Mean (SD) 145.925 (28.231) 152.007 (28.664) 154.173 (26.317)
#> Median 142.705 157.698 155.282
#> Min - Max 85.21 - 195.68 98.90 - 194.62 86.65 - 192.68
#> WEEK 1 DAY 8
#> n 15 15 15 15 15 15
#> Mean (SD) 156.509 (21.097) 10.584 (34.598) 147.480 (33.473) -4.527 (48.895) 143.319 (30.759) -10.854 (34.553)
#> Median 160.711 5.802 155.030 2.758 145.548 -5.636
#> Min - Max 126.84 - 185.53 -53.28 - 91.52 85.22 - 189.88 -77.34 - 90.98 90.37 - 191.58 -65.71 - 49.04
#> WEEK 2 DAY 15
#> n 15 15 15 15 15 15
#> Mean (SD) 144.202 (33.676) -1.723 (27.067) 136.892 (30.178) -15.115 (37.794) 148.622 (27.088) -5.551 (44.670)
#> Median 144.253 5.325 142.679 -14.083 147.102 -11.512
#> Min - Max 62.56 - 203.66 -53.89 - 44.16 70.34 - 174.27 -83.07 - 62.39 108.82 - 200.23 -69.54 - 113.59
#> WEEK 3 DAY 22
#> n 15 15 15 15 15 15
#> Mean (SD) 154.887 (35.374) 8.962 (38.455) 149.761 (28.944) -2.247 (44.835) 150.460 (21.352) -3.712 (37.984)
#> Median 158.938 17.191 155.044 -1.796 156.505 -7.606
#> Min - Max 112.32 - 218.83 -47.28 - 96.18 84.42 - 192.92 -110.20 - 94.02 94.70 - 180.41 -74.91 - 72.74
#> WEEK 4 DAY 29
#> n 15 15 15 15 15 15
#> Mean (SD) 150.159 (32.249) 4.234 (32.965) 156.043 (22.863) 4.036 (42.494) 145.714 (22.980) -8.458 (33.175)
#> Median 145.506 3.754 149.094 -10.000 150.797 -14.432
#> Min - Max 69.37 - 210.43 -89.16 - 54.32 113.57 - 195.10 -71.44 - 77.75 106.91 - 188.09 -41.95 - 65.16
#> WEEK 5 DAY 36
#> n 15 15 15 15 15 15
#> Mean (SD) 155.964 (30.945) 10.039 (42.252) 156.387 (35.274) 4.380 (51.782) 143.592 (33.170) -10.581 (44.799)
#> Median 158.142 1.448 164.552 7.060 148.501 -2.385
#> Min - Max 110.61 - 212.47 -53.91 - 90.45 63.28 - 198.79 -131.34 - 86.84 92.18 - 191.05 -78.77 - 64.353. Absolute Value by Visit
To display only the absolute value, specify
summaryvars = "AVAL".
run(cfbt01, proc_data, dataset = "advs", summaryvars = "AVAL")
#> A: Drug X B: Placebo C: Combination
#> Value at Visit Value at Visit Value at Visit
#> Analysis Visit (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————
#> Diastolic Blood Pressure
#> SCREENING
#> n 15 15 15
#> Mean (SD) 94.385 (17.067) 106.381 (20.586) 106.468 (12.628)
#> Median 94.933 111.133 108.359
#> Min - Max 55.71 - 122.00 60.21 - 131.91 83.29 - 127.17
#> BASELINE
#> n 15 15 15
#> Mean (SD) 96.133 (22.458) 108.111 (15.074) 103.149 (19.752)
#> Median 93.328 108.951 102.849
#> Min - Max 60.58 - 136.59 83.44 - 131.62 66.05 - 136.55
#> WEEK 1 DAY 8
#> n 15 15 15
#> Mean (SD) 98.977 (21.359) 104.110 (16.172) 100.826 (19.027)
#> Median 92.447 107.703 103.058
#> Min - Max 67.55 - 130.37 70.91 - 132.89 70.04 - 128.68
#> WEEK 2 DAY 15
#> n 15 15 15
#> Mean (SD) 99.758 (14.477) 97.473 (17.296) 94.272 (16.961)
#> Median 101.498 99.501 96.789
#> Min - Max 71.98 - 122.81 53.80 - 125.81 63.45 - 117.47
#> WEEK 3 DAY 22
#> n 15 15 15
#> Mean (SD) 99.101 (26.109) 91.984 (16.925) 94.586 (13.560)
#> Median 101.146 91.244 98.398
#> Min - Max 47.68 - 162.22 67.80 - 119.72 73.50 - 115.43
#> WEEK 4 DAY 29
#> n 15 15 15
#> Mean (SD) 103.400 (22.273) 96.467 (19.451) 108.338 (18.417)
#> Median 98.168 97.385 107.555
#> Min - Max 63.09 - 148.25 63.35 - 131.57 68.78 - 132.52
#> WEEK 5 DAY 36
#> n 15 15 15
#> Mean (SD) 93.222 (18.536) 97.890 (20.701) 95.317 (16.401)
#> Median 90.799 99.049 93.876
#> Min - Max 63.55 - 139.11 69.47 - 137.64 71.91 - 138.54
#> Systolic Blood Pressure
#> SCREENING
#> n 15 15 15
#> Mean (SD) 154.073 (33.511) 157.840 (34.393) 152.407 (22.311)
#> Median 156.169 161.670 149.556
#> Min - Max 78.31 - 210.70 79.76 - 210.40 108.21 - 184.88
#> BASELINE
#> n 15 15 15
#> Mean (SD) 145.925 (28.231) 152.007 (28.664) 154.173 (26.317)
#> Median 142.705 157.698 155.282
#> Min - Max 85.21 - 195.68 98.90 - 194.62 86.65 - 192.68
#> WEEK 1 DAY 8
#> n 15 15 15
#> Mean (SD) 156.509 (21.097) 147.480 (33.473) 143.319 (30.759)
#> Median 160.711 155.030 145.548
#> Min - Max 126.84 - 185.53 85.22 - 189.88 90.37 - 191.58
#> WEEK 2 DAY 15
#> n 15 15 15
#> Mean (SD) 144.202 (33.676) 136.892 (30.178) 148.622 (27.088)
#> Median 144.253 142.679 147.102
#> Min - Max 62.56 - 203.66 70.34 - 174.27 108.82 - 200.23
#> WEEK 3 DAY 22
#> n 15 15 15
#> Mean (SD) 154.887 (35.374) 149.761 (28.944) 150.460 (21.352)
#> Median 158.938 155.044 156.505
#> Min - Max 112.32 - 218.83 84.42 - 192.92 94.70 - 180.41
#> WEEK 4 DAY 29
#> n 15 15 15
#> Mean (SD) 150.159 (32.249) 156.043 (22.863) 145.714 (22.980)
#> Median 145.506 149.094 150.797
#> Min - Max 69.37 - 210.43 113.57 - 195.10 106.91 - 188.09
#> WEEK 5 DAY 36
#> n 15 15 15
#> Mean (SD) 155.964 (30.945) 156.387 (35.274) 143.592 (33.170)
#> Median 158.142 164.552 148.501
#> Min - Max 110.61 - 212.47 63.28 - 198.79 92.18 - 191.05
Concomitant Medications by Medication Class and Preferred
Name (CMT01A)
1. Concomitant Medications by Medication Class and Preferred Name
- The
cmt01atemplate displays concomitant medications byATC Level 2and Preferred Name by default. - The template does not include the column of total by default.
- The template sort medication class and preferred name by alphabetical order by default.
run(cmt01a, syn_data)
#> ATC Level 2 Text A: Drug X B: Placebo C: Combination
#> Other Treatment (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one treatment 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of treatments 58 59 99
#> ATCCLAS2 A
#> Total number of patients with at least one treatment 10 (66.7%) 11 (73.3%) 12 (80.0%)
#> Total number of treatments 15 21 28
#> medname A_3/3 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> medname A_2/3 5 (33.3%) 6 (40.0%) 7 (46.7%)
#> medname A_1/3 4 (26.7%) 3 (20.0%) 8 (53.3%)
#> ATCCLAS2 A p2
#> Total number of patients with at least one treatment 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> Total number of treatments 6 8 8
#> medname A_3/3 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> ATCCLAS2 B
#> Total number of patients with at least one treatment 12 (80.0%) 10 (66.7%) 14 (93.3%)
#> Total number of treatments 30 30 52
#> medname B_3/4 8 (53.3%) 6 (40.0%) 8 (53.3%)
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> medname B_4/4 4 (26.7%) 5 (33.3%) 8 (53.3%)
#> ATCCLAS2 B p2
#> Total number of patients with at least one treatment 10 (66.7%) 8 (53.3%) 12 (80.0%)
#> Total number of treatments 18 17 25
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> ATCCLAS2 B p3
#> Total number of patients with at least one treatment 10 (66.7%) 8 (53.3%) 12 (80.0%)
#> Total number of treatments 18 17 25
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> ATCCLAS2 C
#> Total number of patients with at least one treatment 9 (60.0%) 7 (46.7%) 12 (80.0%)
#> Total number of treatments 13 8 19
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname C_1/2 6 (40.0%) 2 (13.3%) 6 (40.0%)
#> ATCCLAS2 C p2
#> Total number of patients with at least one treatment 9 (60.0%) 7 (46.7%) 12 (80.0%)
#> Total number of treatments 13 8 19
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname C_1/2 6 (40.0%) 2 (13.3%) 6 (40.0%)
#> ATCCLAS2 C p3
#> Total number of patients with at least one treatment 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> Total number of treatments 5 5 12
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
2. Concomitant Medications by Medication Class and Preferred
Name (changing ATC class level)
run(cmt01a, syn_data, row_split_var = "ATC1")
#> ATC Level 1 Text A: Drug X B: Placebo C: Combination
#> Other Treatment (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one treatment 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of treatments 58 59 99
#> ATCCLAS1 A
#> Total number of patients with at least one treatment 10 (66.7%) 11 (73.3%) 12 (80.0%)
#> Total number of treatments 15 21 28
#> medname A_3/3 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> medname A_2/3 5 (33.3%) 6 (40.0%) 7 (46.7%)
#> medname A_1/3 4 (26.7%) 3 (20.0%) 8 (53.3%)
#> ATCCLAS1 A p2
#> Total number of patients with at least one treatment 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> Total number of treatments 6 8 8
#> medname A_3/3 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> ATCCLAS1 B
#> Total number of patients with at least one treatment 12 (80.0%) 10 (66.7%) 14 (93.3%)
#> Total number of treatments 30 30 52
#> medname B_3/4 8 (53.3%) 6 (40.0%) 8 (53.3%)
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> medname B_4/4 4 (26.7%) 5 (33.3%) 8 (53.3%)
#> ATCCLAS1 B p2
#> Total number of patients with at least one treatment 10 (66.7%) 8 (53.3%) 12 (80.0%)
#> Total number of treatments 18 17 25
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> ATCCLAS1 B p3
#> Total number of patients with at least one treatment 10 (66.7%) 8 (53.3%) 12 (80.0%)
#> Total number of treatments 18 17 25
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> ATCCLAS1 C
#> Total number of patients with at least one treatment 9 (60.0%) 7 (46.7%) 12 (80.0%)
#> Total number of treatments 13 8 19
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname C_1/2 6 (40.0%) 2 (13.3%) 6 (40.0%)
#> ATCCLAS1 C p2
#> Total number of patients with at least one treatment 9 (60.0%) 7 (46.7%) 12 (80.0%)
#> Total number of treatments 13 8 19
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname C_1/2 6 (40.0%) 2 (13.3%) 6 (40.0%)
#> ATCCLAS1 C p3
#> Total number of patients with at least one treatment 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> Total number of treatments 5 5 12
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)3. Concomitant Medications by Medication Class and Preferred Name (classes sorted by frequency)
The argument sort_by_freq = TRUE sort medication class
by frequency.
run(cmt01a, syn_data, sort_by_freq = TRUE)
#> ATC Level 2 Text A: Drug X B: Placebo C: Combination
#> Other Treatment (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one treatment 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of treatments 58 59 99
#> ATCCLAS2 B
#> Total number of patients with at least one treatment 12 (80.0%) 10 (66.7%) 14 (93.3%)
#> Total number of treatments 30 30 52
#> medname B_3/4 8 (53.3%) 6 (40.0%) 8 (53.3%)
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> medname B_4/4 4 (26.7%) 5 (33.3%) 8 (53.3%)
#> ATCCLAS2 A
#> Total number of patients with at least one treatment 10 (66.7%) 11 (73.3%) 12 (80.0%)
#> Total number of treatments 15 21 28
#> medname A_3/3 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> medname A_2/3 5 (33.3%) 6 (40.0%) 7 (46.7%)
#> medname A_1/3 4 (26.7%) 3 (20.0%) 8 (53.3%)
#> ATCCLAS2 B p2
#> Total number of patients with at least one treatment 10 (66.7%) 8 (53.3%) 12 (80.0%)
#> Total number of treatments 18 17 25
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> ATCCLAS2 B p3
#> Total number of patients with at least one treatment 10 (66.7%) 8 (53.3%) 12 (80.0%)
#> Total number of treatments 18 17 25
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> ATCCLAS2 C
#> Total number of patients with at least one treatment 9 (60.0%) 7 (46.7%) 12 (80.0%)
#> Total number of treatments 13 8 19
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname C_1/2 6 (40.0%) 2 (13.3%) 6 (40.0%)
#> ATCCLAS2 C p2
#> Total number of patients with at least one treatment 9 (60.0%) 7 (46.7%) 12 (80.0%)
#> Total number of treatments 13 8 19
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname C_1/2 6 (40.0%) 2 (13.3%) 6 (40.0%)
#> ATCCLAS2 A p2
#> Total number of patients with at least one treatment 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> Total number of treatments 6 8 8
#> medname A_3/3 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> ATCCLAS2 C p3
#> Total number of patients with at least one treatment 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> Total number of treatments 5 5 12
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)4. Concomitant Medications by Medication Class and Preferred Name (total number of treatments per medication class suppressed)
The cmt01a template includes the
analysis of ‘total number of treatments’ by default, modify the argument
summary_labels to change it.
run(cmt01a, syn_data, summary_labels = list(TOTAL = cmt01_label, ATC2 = cmt01_label[1]))
#> ATC Level 2 Text A: Drug X B: Placebo C: Combination
#> Other Treatment (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one treatment 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of treatments 58 59 99
#> ATCCLAS2 A
#> Total number of patients with at least one treatment 10 (66.7%) 11 (73.3%) 12 (80.0%)
#> medname A_3/3 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> medname A_2/3 5 (33.3%) 6 (40.0%) 7 (46.7%)
#> medname A_1/3 4 (26.7%) 3 (20.0%) 8 (53.3%)
#> ATCCLAS2 A p2
#> Total number of patients with at least one treatment 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> medname A_3/3 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> ATCCLAS2 B
#> Total number of patients with at least one treatment 12 (80.0%) 10 (66.7%) 14 (93.3%)
#> medname B_3/4 8 (53.3%) 6 (40.0%) 8 (53.3%)
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> medname B_4/4 4 (26.7%) 5 (33.3%) 8 (53.3%)
#> ATCCLAS2 B p2
#> Total number of patients with at least one treatment 10 (66.7%) 8 (53.3%) 12 (80.0%)
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> ATCCLAS2 B p3
#> Total number of patients with at least one treatment 10 (66.7%) 8 (53.3%) 12 (80.0%)
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> ATCCLAS2 C
#> Total number of patients with at least one treatment 9 (60.0%) 7 (46.7%) 12 (80.0%)
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname C_1/2 6 (40.0%) 2 (13.3%) 6 (40.0%)
#> ATCCLAS2 C p2
#> Total number of patients with at least one treatment 9 (60.0%) 7 (46.7%) 12 (80.0%)
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname C_1/2 6 (40.0%) 2 (13.3%) 6 (40.0%)
#> ATCCLAS2 C p3
#> Total number of patients with at least one treatment 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
Concomitant Medications by Preferred Name
(CMT02_PT)
1. Concomitant Medications by Preferred Name
- The
cmt02_pttemplate displays concomitant medications by Preferred Name by default. - The template does not include the column of total by default.
- The template sorts preferred name by alphabetical order by default.
Set the argument
sort_by_freq = TRUEto sort preferred names by frequency.
run(cmt02_pt, syn_data)
#> A: Drug X B: Placebo C: Combination
#> Other Treatment (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one treatment 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of treatments 58 59 99
#> medname B_3/4 8 (53.3%) 6 (40.0%) 8 (53.3%)
#> medname B_2/4 6 (40.0%) 5 (33.3%) 10 (66.7%)
#> medname A_3/3 5 (33.3%) 8 (53.3%) 6 (40.0%)
#> medname B_1/4 7 (46.7%) 6 (40.0%) 6 (40.0%)
#> medname A_2/3 5 (33.3%) 6 (40.0%) 7 (46.7%)
#> medname B_4/4 4 (26.7%) 5 (33.3%) 8 (53.3%)
#> medname C_2/2 4 (26.7%) 5 (33.3%) 7 (46.7%)
#> medname A_1/3 4 (26.7%) 3 (20.0%) 8 (53.3%)
#> medname C_1/2 6 (40.0%) 2 (13.3%) 6 (40.0%)
Cox Regression (COXT01)
1. Cox Regression
- The
coxt01template produces the standard Cox regression output. - Users are expected to pre-process the input analysis data by selecting a time-to-event parameter to be analyzed. The example below is based on the time-to-event parameter “Duration of Confirmed Response by Investigator”.
- The time variable in the model is specified through the
time_varargument. By default,time_varis set to"AVAL", which comes fromADTTE.AVAL. - The event variable in the model is specified through the
event_varargument. By default,event_varis set to"EVENT", which is derived based on the censoring indicatorADTTE.CNSRin the pre-processing functioncoxt01_pre. - If there are more than two treatment groups present in the input
analysis data, users are also expected to select only two treatment
groups. The example below is based on treatment groups
"Arm A"and"Arm B".
proc_data <- log_filter(syn_data, PARAMCD == "OS", "adtte")
proc_data <- log_filter(proc_data, ARMCD != "ARM C", "adsl")
run(coxt01, proc_data, time_var = "AVAL", event_var = "EVENT")
#> Treatment Effect Adjusted for Covariate
#> Effect/Covariate Included in the Model n Hazard Ratio 95% CI p-value
#> —————————————————————————————————————————————————————————————————————————————————————————
#> Treatment:
#> B: Placebo vs control (A: Drug X) 30 2.71 (0.93, 7.88) 0.0666
#> Covariate:
#> Sex 30 2.91 (0.97, 8.73) 0.0567
#> RACE 30 3.09 (1.01, 9.50) 0.0487
#> Age (yr) 30 2.89 (0.97, 8.59) 0.05662. Cox Regression (with interaction term)
To add the interaction term to the model,
interaction = TRUE, which is passed to
tern::control_coxreg(), needs to be specified.
run(coxt01, proc_data, covariates = "AAGE", interaction = TRUE)
#> Treatment Effect Adjusted for Covariate
#> Effect/Covariate Included in the Model n Hazard Ratio 95% CI p-value Interaction p-value
#> —————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Treatment:
#> B: Placebo vs control (A: Drug X) 30 2.71 (0.93, 7.88) 0.0666
#> Covariate:
#> Age (yr) 30 0.3666
#> 32 2.87 (0.98, 8.41)3. Cox Regression (specifying covariates)
- By default,
"SEX","RACE"and"AAGE"are used as the covariates for the model. - Users can specify a different set of covariates through the
covariatesargument. In the example below,"RACE"and"AAGE"are used as covariates.
run(coxt01, proc_data, covariates = c("RACE", "AAGE"))
#> Treatment Effect Adjusted for Covariate
#> Effect/Covariate Included in the Model n Hazard Ratio 95% CI p-value
#> —————————————————————————————————————————————————————————————————————————————————————————
#> Treatment:
#> B: Placebo vs control (A: Drug X) 30 2.71 (0.93, 7.88) 0.0666
#> Covariate:
#> RACE 30 3.09 (1.01, 9.50) 0.0487
#> Age (yr) 30 2.89 (0.97, 8.59) 0.05664. Cox Regression (setting strata, ties, and alpha level)
- By default,
strata = NULL(no stratification),ties = "exact"(equivalent toDISCRETEin SAS), andconf_level = 0.95are applied. - Users can specify one or more stratification variables via the
strataargument. - Other tie handling methods, i.e.,
"efron"or"breslow", can be specified in thetieargument, which is passed totern::control_coxreg(). - Users can also customize the alpha level for the confidence
intervals through the
conf_levelargument, which is passed totern::control_coxreg().
run(coxt01, proc_data, covariates = c("SEX", "AAGE"), strata = c("RACE"), conf_level = 0.90)
#> Treatment Effect Adjusted for Covariate
#> Effect/Covariate Included in the Model n Hazard Ratio 90% CI p-value
#> —————————————————————————————————————————————————————————————————————————————————————————
#> Treatment:
#> B: Placebo vs control (A: Drug X) 30 2.69 (1.07, 6.76) 0.0785
#> Covariate:
#> Sex 30 2.90 (1.12, 7.54) 0.0668
#> Age (yr) 30 2.72 (1.08, 6.85) 0.0755
Multi-variable Cox Regression
(COXT02)
1. Multi-variable Cox Regression
- The
coxt02template produces the standard multi-variable cox regression output. - Users are expected to pre-process the input analysis data by selecting a time-to-event parameter to be analyzed. The example below is based on the time-to-event parameter “Duration of Confirmed Response by Investigator”.
- The time variable in the model is specified through the
time_varargument. By default,time_varis set to"AVAL", which comes fromADTTE.AVAL. - The event variable in the model is specified through the
event_varargument. By default,event_varis set to"EVENT", which is derived based on the censoring indicatorADTTE.CNSRin the pre-processing functioncoxt01_pre.
proc_data <- log_filter(syn_data, PARAMCD == "OS", "adtte")
run(coxt02, proc_data, time_var = "AVAL", event_var = "EVENT")
#> Effect/Covariate Included in the Model Hazard Ratio 95% CI p-value
#> —————————————————————————————————————————————————————————————————————————————————————————————
#> Treatment:
#> Description of Planned Arm (reference = A: Drug X) 0.1630
#> B: Placebo 2.92 (0.93, 9.17) 0.0672
#> C: Combination 1.56 (0.47, 5.10) 0.4659
#> Covariate:
#> Sex (reference = F)
#> M 1.03 (0.41, 2.55) 0.9549
#> RACE (reference = AMERICAN INDIAN OR ALASKA NATIVE) 0.8498
#> ASIAN 1.22 (0.27, 5.55) 0.7967
#> BLACK OR AFRICAN AMERICAN 0.81 (0.12, 5.70) 0.8340
#> WHITE 1.57 (0.26, 9.67) 0.6258
#> Age (yr)
#> All 0.99 (0.93, 1.05) 0.66502. Multi-variable Cox Regression (specifying covariates)
- By default,
"SEX","RACE"and"AAGE"are used as the covariates for the model. - Users can specify a different set of covariates through the
covariatesargument. In the example below,"RACE"and"AAGE"are used as covariates.
run(coxt02, proc_data, covariates = c("RACE", "AAGE"))
#> Effect/Covariate Included in the Model Hazard Ratio 95% CI p-value
#> —————————————————————————————————————————————————————————————————————————————————————————————
#> Treatment:
#> Description of Planned Arm (reference = A: Drug X) 0.1390
#> B: Placebo 2.94 (0.97, 8.92) 0.0570
#> C: Combination 1.56 (0.48, 5.09) 0.4605
#> Covariate:
#> RACE (reference = AMERICAN INDIAN OR ALASKA NATIVE) 0.8504
#> ASIAN 1.22 (0.27, 5.54) 0.7972
#> BLACK OR AFRICAN AMERICAN 0.81 (0.12, 5.65) 0.8306
#> WHITE 1.56 (0.26, 9.53) 0.6279
#> Age (yr)
#> All 0.99 (0.93, 1.05) 0.66333. Multi-variable Cox Regression (setting strata, ties, and alpha level)
- By default,
strata = NULL(no stratification),ties = "exact"(equivalent toDISCRETEin SAS), andconf_level = 0.95are applied. - Users can specify one or more stratification variables via the
strataargument. - Other tie handling methods, i.e.,
"efron"or"breslow", can be specified in thetieargument, which is passed totern::control_coxreg(). - Users can also customize the alpha level for the confidence
intervals through the
conf_levelargument, which is passed totern::control_coxreg().
run(coxt02, proc_data, covariates = c("SEX", "AAGE"), strata = c("RACE"), conf_level = 0.90, ties = "efron")
#> Effect/Covariate Included in the Model Hazard Ratio 90% CI p-value
#> ————————————————————————————————————————————————————————————————————————————————————————————
#> Treatment:
#> Description of Planned Arm (reference = A: Drug X) 0.1680
#> B: Placebo 2.85 (1.09, 7.46) 0.0743
#> C: Combination 1.47 (0.54, 4.02) 0.5254
#> Covariate:
#> Sex (reference = F)
#> M 0.98 (0.45, 2.13) 0.9700
#> Age (yr)
#> All 0.99 (0.94, 1.04) 0.6571
Demographics and Baseline Characteristics
(DMT01)
1. Demographics and Baseline Characteristics with All Patients
- The
dmt01template produces the standard demographics and baseline characteristics summary. - This template includes the column of total by default.
run(dmt01, syn_data)
#> A: Drug X B: Placebo C: Combination All Patients
#> (N=15) (N=15) (N=15) (N=45)
#> ————————————————————————————————————————————————————————————————————————————————————————————
#> Age (yr)
#> n 15 15 15 45
#> Mean (SD) 31.3 (5.3) 35.1 (9.0) 36.6 (6.4) 34.3 (7.3)
#> Median 31.0 35.0 35.0 34.0
#> Min - Max 24 - 40 24 - 57 24 - 49 24 - 57
#> Age Group
#> n 15 15 15 45
#> <65 15 (100%) 15 (100%) 15 (100%) 45 (100%)
#> Sex
#> n 15 15 15 45
#> Male 3 (20.0%) 7 (46.7%) 5 (33.3%) 15 (33.3%)
#> Female 12 (80.0%) 8 (53.3%) 10 (66.7%) 30 (66.7%)
#> Ethnicity
#> n 15 15 15 45
#> HISPANIC OR LATINO 2 (13.3%) 0 0 2 (4.4%)
#> NOT HISPANIC OR LATINO 13 (86.7%) 15 (100%) 13 (86.7%) 41 (91.1%)
#> NOT REPORTED 0 0 2 (13.3%) 2 (4.4%)
#> RACE
#> n 15 15 15 45
#> AMERICAN INDIAN OR ALASKA NATIVE 0 2 (13.3%) 1 (6.7%) 3 (6.7%)
#> ASIAN 8 (53.3%) 10 (66.7%) 8 (53.3%) 26 (57.8%)
#> BLACK OR AFRICAN AMERICAN 4 (26.7%) 1 (6.7%) 4 (26.7%) 9 (20.0%)
#> WHITE 3 (20.0%) 2 (13.3%) 2 (13.3%) 7 (15.6%)2. Demographics and Baseline Characteristics without All Patients
To remove the column of total, set the argument
lbl_overall to NULL.
run(dmt01, syn_data, lbl_overall = NULL)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————
#> Age (yr)
#> n 15 15 15
#> Mean (SD) 31.3 (5.3) 35.1 (9.0) 36.6 (6.4)
#> Median 31.0 35.0 35.0
#> Min - Max 24 - 40 24 - 57 24 - 49
#> Age Group
#> n 15 15 15
#> <65 15 (100%) 15 (100%) 15 (100%)
#> Sex
#> n 15 15 15
#> Male 3 (20.0%) 7 (46.7%) 5 (33.3%)
#> Female 12 (80.0%) 8 (53.3%) 10 (66.7%)
#> Ethnicity
#> n 15 15 15
#> HISPANIC OR LATINO 2 (13.3%) 0 0
#> NOT HISPANIC OR LATINO 13 (86.7%) 15 (100%) 13 (86.7%)
#> NOT REPORTED 0 0 2 (13.3%)
#> RACE
#> n 15 15 15
#> AMERICAN INDIAN OR ALASKA NATIVE 0 2 (13.3%) 1 (6.7%)
#> ASIAN 8 (53.3%) 10 (66.7%) 8 (53.3%)
#> BLACK OR AFRICAN AMERICAN 4 (26.7%) 1 (6.7%) 4 (26.7%)
#> WHITE 3 (20.0%) 2 (13.3%) 2 (13.3%)3. Demographics and Baseline Characteristics with an additional study specific continuous variable
- Study specific continuous variables can be added to the standard
demographics and baseline characteristics summary by editing the
argument
summaryvars. To add or remove analyses, you need to pass all variables you would like to include to the argument. - CHEVRON performs the analysis based on the type of variable as defined in the input data.
run(dmt01, syn_data, summaryvars = c("AGE", "AGEGR1", "SEX", "ETHNIC", "RACE", "BBMISI"), lbl_overall = NULL)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————
#> Age
#> n 15 15 15
#> Mean (SD) 31.3 (5.3) 35.1 (9.0) 36.6 (6.4)
#> Median 31.0 35.0 35.0
#> Min - Max 24 - 40 24 - 57 24 - 49
#> Age Group
#> n 15 15 15
#> <65 15 (100%) 15 (100%) 15 (100%)
#> Sex
#> n 15 15 15
#> Male 3 (20.0%) 7 (46.7%) 5 (33.3%)
#> Female 12 (80.0%) 8 (53.3%) 10 (66.7%)
#> Ethnicity
#> n 15 15 15
#> HISPANIC OR LATINO 2 (13.3%) 0 0
#> NOT HISPANIC OR LATINO 13 (86.7%) 15 (100%) 13 (86.7%)
#> NOT REPORTED 0 0 2 (13.3%)
#> RACE
#> n 15 15 15
#> AMERICAN INDIAN OR ALASKA NATIVE 0 2 (13.3%) 1 (6.7%)
#> ASIAN 8 (53.3%) 10 (66.7%) 8 (53.3%)
#> BLACK OR AFRICAN AMERICAN 4 (26.7%) 1 (6.7%) 4 (26.7%)
#> WHITE 3 (20.0%) 2 (13.3%) 2 (13.3%)
#> Baseline BMI
#> n 15 15 15
#> Mean (SD) 29.75 (15.10) 41.08 (26.65) 33.90 (15.39)
#> Median 37.00 33.70 37.80
#> Min - Max 6.4 - 47.9 5.3 - 117.9 -3.5 - 59.04. Demographics and Baseline Characteristics with an additional study specific categorical variable
- Study specific categorical variables can be added to the standard
demographics and baseline characteristics summary by editing the
argument
summaryvars. - To display the values within a categorical variable in pre-specified order, the categorical variable need to be factorized with pre-specified order provided as levels.
proc_data <- syn_data
proc_data$adsl <- proc_data$adsl %>%
mutate(
SEX = reformat(.data$SEX, rule(Male = "M", Female = "F")),
BBMIGR1 = factor(case_when(
BBMISI < 15 ~ "Very severely underweight",
BBMISI >= 15 & BBMISI < 16 ~ "Severely underweight",
BBMISI >= 16 & BBMISI < 18.5 ~ "Underweight",
BBMISI >= 18.5 & BBMISI < 25 ~ "Normal (healthy weight)",
BBMISI >= 25 & BBMISI < 30 ~ "Overweight",
BBMISI >= 30 & BBMISI < 35 ~ "Obese Class I (Moderately obese)",
BBMISI >= 35 & BBMISI < 40 ~ "Obese Class II (Severely obese)",
BBMISI >= 40 ~ "Obese Class III (Very severely obese)"
), levels = c(
"Very severely underweight",
"Severely underweight",
"Underweight",
"Normal (healthy weight)",
"Overweight",
"Obese Class I (Moderately obese)",
"Obese Class II (Severely obese)",
"Obese Class III (Very severely obese)"
))
)
run(dmt01, proc_data, summaryvars = c("AGE", "AGEGR1", "SEX", "ETHNIC", "RACE", "BBMIGR1"), auto_pre = FALSE)
#> A: Drug X B: Placebo C: Combination All Patients
#> (N=15) (N=15) (N=15) (N=45)
#> —————————————————————————————————————————————————————————————————————————————————————————————————
#> Age
#> n 15 15 15 45
#> Mean (SD) 31.3 (5.3) 35.1 (9.0) 36.6 (6.4) 34.3 (7.3)
#> Median 31.0 35.0 35.0 34.0
#> Min - Max 24 - 40 24 - 57 24 - 49 24 - 57
#> Age Group
#> n 15 15 15 45
#> <65 15 (100%) 15 (100%) 15 (100%) 45 (100%)
#> Sex
#> n 15 15 15 45
#> Male 3 (20.0%) 7 (46.7%) 5 (33.3%) 15 (33.3%)
#> Female 12 (80.0%) 8 (53.3%) 10 (66.7%) 30 (66.7%)
#> Ethnicity
#> n 15 15 15 45
#> HISPANIC OR LATINO 2 (13.3%) 0 0 2 (4.4%)
#> NOT HISPANIC OR LATINO 13 (86.7%) 15 (100%) 13 (86.7%) 41 (91.1%)
#> NOT REPORTED 0 0 2 (13.3%) 2 (4.4%)
#> RACE
#> n 15 15 15 45
#> AMERICAN INDIAN OR ALASKA NATIVE 0 2 (13.3%) 1 (6.7%) 3 (6.7%)
#> ASIAN 8 (53.3%) 10 (66.7%) 8 (53.3%) 26 (57.8%)
#> BLACK OR AFRICAN AMERICAN 4 (26.7%) 1 (6.7%) 4 (26.7%) 9 (20.0%)
#> WHITE 3 (20.0%) 2 (13.3%) 2 (13.3%) 7 (15.6%)
#> BBMIGR1
#> n 15 15 15 45
#> Very severely underweight 4 (26.7%) 1 (6.7%) 1 (6.7%) 6 (13.3%)
#> Underweight 1 (6.7%) 0 0 1 (2.2%)
#> Normal (healthy weight) 1 (6.7%) 3 (20.0%) 4 (26.7%) 8 (17.8%)
#> Overweight 0 1 (6.7%) 1 (6.7%) 2 (4.4%)
#> Obese Class I (Moderately obese) 0 3 (20.0%) 0 3 (6.7%)
#> Obese Class II (Severely obese) 4 (26.7%) 1 (6.7%) 3 (20.0%) 8 (17.8%)
#> Obese Class III (Very severely obese) 5 (33.3%) 6 (40.0%) 6 (40.0%) 17 (37.8%)
5. Demographics and Baseline Characteristics with additional
vital signs baseline values from ADVS or
ADSUB
To add baseline vital signs or other baseline characteristics to the
demographics and baseline characteristics summary, manual preprocess of
input adsl dataset is expected and merge the vital signs
baseline values from advs (where
ADVS.ABLFL == "Y") or adsub with
adsl by unique subject identifier.
proc_data <- syn_data
diabpbl <- proc_data$advs %>%
filter(ABLFL == "Y" & PARAMCD == "DIABP") %>%
mutate(DIABPBL = AVAL) %>%
select("STUDYID", "USUBJID", "DIABPBL")
proc_data$adsl <- proc_data$adsl %>%
mutate(SEX = reformat(.data$SEX, rule(Male = "M", Female = "F"))) %>%
left_join(diabpbl, by = c("STUDYID", "USUBJID"))
run(dmt01, proc_data, summaryvars = c("AGE", "AGEGR1", "SEX", "ETHNIC", "RACE", "DIABPBL"), auto_pre = FALSE)
#> A: Drug X B: Placebo C: Combination All Patients
#> (N=15) (N=15) (N=15) (N=45)
#> —————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Age
#> n 15 15 15 45
#> Mean (SD) 31.3 (5.3) 35.1 (9.0) 36.6 (6.4) 34.3 (7.3)
#> Median 31.0 35.0 35.0 34.0
#> Min - Max 24 - 40 24 - 57 24 - 49 24 - 57
#> Age Group
#> n 15 15 15 45
#> <65 15 (100%) 15 (100%) 15 (100%) 45 (100%)
#> Sex
#> n 15 15 15 45
#> Male 3 (20.0%) 7 (46.7%) 5 (33.3%) 15 (33.3%)
#> Female 12 (80.0%) 8 (53.3%) 10 (66.7%) 30 (66.7%)
#> Ethnicity
#> n 15 15 15 45
#> HISPANIC OR LATINO 2 (13.3%) 0 0 2 (4.4%)
#> NOT HISPANIC OR LATINO 13 (86.7%) 15 (100%) 13 (86.7%) 41 (91.1%)
#> NOT REPORTED 0 0 2 (13.3%) 2 (4.4%)
#> RACE
#> n 15 15 15 45
#> AMERICAN INDIAN OR ALASKA NATIVE 0 2 (13.3%) 1 (6.7%) 3 (6.7%)
#> ASIAN 8 (53.3%) 10 (66.7%) 8 (53.3%) 26 (57.8%)
#> BLACK OR AFRICAN AMERICAN 4 (26.7%) 1 (6.7%) 4 (26.7%) 9 (20.0%)
#> WHITE 3 (20.0%) 2 (13.3%) 2 (13.3%) 7 (15.6%)
#> Analysis Value
#> n 15 15 15 45
#> Mean (SD) 96.132511 (22.458204) 108.110944 (15.074451) 103.148818 (19.751687) 102.464091 (19.534945)
#> Median 93.328321 108.951358 102.849019 102.396129
#> Min - Max 60.58490 - 136.59343 83.44277 - 131.61501 66.05223 - 136.55256 60.58490 - 136.59343
Patient Disposition (DST01)
1. Patient Disposition
- The
dst01template produces the standard patient disposition summary. - The template includes the column of total by default. Use
lbl_overall = NULLto suppress the default.
run(dst01, syn_data, lbl_overall = NULL)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ——————————————————————————————————————————————————————————————————————————
#> Completed 10 (66.7%) 10 (66.7%) 10 (66.7%)
#> Discontinued 5 (33.3%) 5 (33.3%) 5 (33.3%)
#> ADVERSE EVENT 0 0 1 (6.7%)
#> DEATH 2 (13.3%) 4 (26.7%) 3 (20.0%)
#> LACK OF EFFICACY 2 (13.3%) 0 0
#> PHYSICIAN DECISION 0 0 1 (6.7%)
#> PROTOCOL VIOLATION 0 1 (6.7%) 0
#> WITHDRAWAL BY PARENT/GUARDIAN 1 (6.7%) 0 02. Patient Disposition (with grouping of reasons)
- The syntax below produces the standard patient disposition summary with grouping of the discontinuation reasons.
- The variable [
ADSL.DCSREASGP] that groups the discontinuation reasons needs to be derived manually and provided in the inputadsldataset.
run(dst01, syn_data, detail_vars = list(Discontinued = c("DCSREASGP", "DCSREAS")), lbl_overall = NULL)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————
#> Completed 10 (66.7%) 10 (66.7%) 10 (66.7%)
#> Discontinued 5 (33.3%) 5 (33.3%) 5 (33.3%)
#> Safety
#> ADVERSE EVENT 0 0 1 (6.7%)
#> DEATH 2 (13.3%) 4 (26.7%) 3 (20.0%)
#> Non-Safety
#> LACK OF EFFICACY 2 (13.3%) 0 0
#> PHYSICIAN DECISION 0 0 1 (6.7%)
#> PROTOCOL VIOLATION 0 1 (6.7%) 0
#> WITHDRAWAL BY PARENT/GUARDIAN 1 (6.7%) 0 03. Patient Disposition (adding end of treatment status)
The syntax below adds the end of treatment status to the standard
patient disposition summary by providing the end of treatment status
variable to the argument trt_status_var.
run(dst01, syn_data, trt_status_var = "EOTSTT", lbl_overall = NULL)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ——————————————————————————————————————————————————————————————————————————
#> Completed 10 (66.7%) 10 (66.7%) 10 (66.7%)
#> Discontinued 5 (33.3%) 5 (33.3%) 5 (33.3%)
#> ADVERSE EVENT 0 0 1 (6.7%)
#> DEATH 2 (13.3%) 4 (26.7%) 3 (20.0%)
#> LACK OF EFFICACY 2 (13.3%) 0 0
#> PHYSICIAN DECISION 0 0 1 (6.7%)
#> PROTOCOL VIOLATION 0 1 (6.7%) 0
#> WITHDRAWAL BY PARENT/GUARDIAN 1 (6.7%) 0 0
#> Completed Treatment 8 (53.3%) 4 (26.7%) 5 (33.3%)
#> Ongoing Treatment 4 (26.7%) 6 (40.0%) 4 (26.7%)
#> Discontinued Treatment 3 (20.0%) 5 (33.3%) 6 (40.0%)4. Patient Disposition (adding details of study ongoing status)
The syntax adds the details of study ongoing/alive status to the
standard patient disposition summary by modifying the argument
detail_vars.
run(dst01, syn_data, detail_vars = list(Discontinued = "DCSREAS", Ongoing = "STDONS"))
#> A: Drug X B: Placebo C: Combination All Patients
#> (N=15) (N=15) (N=15) (N=45)
#> —————————————————————————————————————————————————————————————————————————————————————————
#> Completed 10 (66.7%) 10 (66.7%) 10 (66.7%) 30 (66.7%)
#> Discontinued 5 (33.3%) 5 (33.3%) 5 (33.3%) 15 (33.3%)
#> ADVERSE EVENT 0 0 1 (6.7%) 1 (2.2%)
#> DEATH 2 (13.3%) 4 (26.7%) 3 (20.0%) 9 (20.0%)
#> LACK OF EFFICACY 2 (13.3%) 0 0 2 (4.4%)
#> PHYSICIAN DECISION 0 0 1 (6.7%) 1 (2.2%)
#> PROTOCOL VIOLATION 0 1 (6.7%) 0 1 (2.2%)
#> WITHDRAWAL BY PARENT/GUARDIAN 1 (6.7%) 0 0 1 (2.2%)
Deaths (DTHT01)
1. Deaths
The dtht01 template produces the
standard deaths output.
run(dst01, syn_data)
#> A: Drug X B: Placebo C: Combination All Patients
#> (N=15) (N=15) (N=15) (N=45)
#> —————————————————————————————————————————————————————————————————————————————————————————
#> Completed 10 (66.7%) 10 (66.7%) 10 (66.7%) 30 (66.7%)
#> Discontinued 5 (33.3%) 5 (33.3%) 5 (33.3%) 15 (33.3%)
#> ADVERSE EVENT 0 0 1 (6.7%) 1 (2.2%)
#> DEATH 2 (13.3%) 4 (26.7%) 3 (20.0%) 9 (20.0%)
#> LACK OF EFFICACY 2 (13.3%) 0 0 2 (4.4%)
#> PHYSICIAN DECISION 0 0 1 (6.7%) 1 (2.2%)
#> PROTOCOL VIOLATION 0 1 (6.7%) 0 1 (2.2%)
#> WITHDRAWAL BY PARENT/GUARDIAN 1 (6.7%) 0 0 1 (2.2%)2. Deaths (adding “Primary Cause of Death” details for ‘Other’ category)
run(dtht01, syn_data, other_category = TRUE)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————
#> Total number of deaths 2 (13.3%) 4 (26.7%) 3 (20.0%)
#> Primary Cause of Death
#> n 2 4 3
#> Adverse Event 1 (50.0%) 2 (50.0%) 1 (33.3%)
#> Progressive Disease 1 (50.0%) 0 2 (66.7%)
#> Other 0 2 (50.0%) 0
#> LOST TO FOLLOW UP 0 1 (50%) 0
#> SUICIDE 0 1 (50%) 0NOTE: In order to avoid the warning above and display ‘Other’ as the
last category under “Primary Cause of Death” right above the detailed
reasons for “Other”, the user is expected to manually provide levels to
ADSL.DTHCAT based on categories available in the
dataset.
3. Deaths (adding summary by days from last study drug administration)
Setting time_since_last_dose to TRUE, the
syntax produces the count of deaths by days from last study drug
administration as well as the count of deaths by primary cause and days
from last study drug administration.
run(dtht01, syn_data, time_since_last_dose = TRUE)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of deaths 2 (13.3%) 4 (26.7%) 3 (20.0%)
#> Days from last drug administration
#> n 2 4 3
#> <=30 2 (100%) 1 (25.0%) 2 (66.7%)
#> >30 0 3 (75.0%) 1 (33.3%)
#> Primary cause by days from last study drug administration
#> <=30
#> n 2 1 2
#> Adverse Event 1 (50.0%) 0 1 (50.0%)
#> Progressive Disease 1 (50.0%) 0 1 (50.0%)
#> Other 0 1 (100%) 0
#> >30
#> n 0 3 1
#> Adverse Event 0 2 (66.7%) 0
#> Progressive Disease 0 0 1 (100%)
#> Other 0 1 (33.3%) 0
ECG Results and Change from Baseline by Visit
(EGT01)
1. ECG Results and Change from Baseline by Visit
The egt01 template produces the
standard ECG results and change from baseline by visit summary.
run(egt01, syn_data)
#> A: Drug X B: Placebo C: Combination
#> Change from Change from Change from
#> Value at Visit Baseline Value at Visit Baseline Value at Visit Baseline
#> Analysis Visit (N=15) (N=15) (N=15) (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Heart Rate
#> BASELINE
#> n 15 15 15
#> Mean (SD) 76.594 (17.889) 69.899 (18.788) 70.492 (18.175)
#> Median 77.531 77.174 74.111
#> Min - Max 46.50 - 106.68 26.42 - 97.69 45.37 - 115.49
#> WEEK 1 DAY 8
#> n 15 15 15 15 15 15
#> Mean (SD) 71.140 (23.441) -5.454 (25.128) 70.958 (14.877) 1.059 (23.345) 67.450 (18.932) -3.043 (23.753)
#> Median 77.210 -2.152 70.033 -8.403 68.471 0.181
#> Min - Max 8.53 - 102.63 -50.97 - 36.54 44.85 - 93.79 -25.34 - 60.50 38.90 - 100.05 -52.20 - 33.13
#> WEEK 2 DAY 15
#> n 15 15 15 15 15 15
#> Mean (SD) 69.350 (16.083) -7.244 (28.960) 76.096 (14.958) 6.198 (29.319) 63.694 (12.920) -6.799 (23.949)
#> Median 65.746 -11.369 75.323 0.255 61.076 -4.954
#> Min - Max 47.22 - 101.44 -49.59 - 42.91 47.50 - 111.40 -37.51 - 69.34 43.25 - 86.13 -52.70 - 40.76
#> WEEK 3 DAY 22
#> n 15 15 15 15 15 15
#> Mean (SD) 73.894 (24.576) -2.700 (32.079) 67.635 (19.114) -2.263 (29.989) 72.054 (19.308) 1.562 (27.494)
#> Median 69.296 5.492 68.468 -2.093 68.686 -5.848
#> Min - Max 44.15 - 131.73 -62.53 - 38.19 31.89 - 108.87 -52.26 - 66.81 32.16 - 109.86 -49.61 - 35.23
#> WEEK 4 DAY 29
#> n 15 15 15 15 15 15
#> Mean (SD) 73.241 (19.256) -3.353 (29.170) 66.524 (25.487) -3.374 (36.024) 66.600 (22.839) -3.892 (24.140)
#> Median 68.689 0.232 66.397 -11.730 64.969 -6.827
#> Min - Max 33.71 - 111.54 -55.14 - 65.04 19.66 - 111.29 -60.39 - 61.00 10.35 - 100.88 -50.72 - 26.77
#> WEEK 5 DAY 36
#> n 15 15 15 15 15 15
#> Mean (SD) 61.690 (22.182) -14.904 (30.330) 60.712 (20.025) -9.187 (24.587) 72.683 (23.495) 2.191 (26.654)
#> Median 57.925 -12.660 60.454 -16.100 77.585 14.635
#> Min - Max 23.89 - 103.74 -60.00 - 57.24 32.53 - 102.02 -52.56 - 50.96 31.21 - 105.05 -42.90 - 34.64
#> QT Duration
#> BASELINE
#> n 15 15 15
#> Mean (SD) 335.294 (123.231) 363.104 (68.160) 347.311 (86.236)
#> Median 372.731 386.316 348.254
#> Min - Max 121.28 - 554.97 214.65 - 445.53 170.80 - 508.54
#> WEEK 1 DAY 8
#> n 15 15 15 15 15 15
#> Mean (SD) 357.361 (85.688) 22.067 (144.166) 415.225 (105.425) 52.121 (144.259) 321.078 (107.553) -26.233 (129.135)
#> Median 344.797 49.432 421.950 62.762 307.962 -17.006
#> Min - Max 241.22 - 517.39 -207.23 - 245.36 234.11 - 604.72 -190.70 - 364.94 118.36 - 480.29 -363.11 - 163.67
#> WEEK 2 DAY 15
#> n 15 15 15 15 15 15
#> Mean (SD) 344.883 (106.793) 9.589 (174.797) 370.548 (80.862) 7.444 (91.301) 354.129 (95.133) 6.818 (142.397)
#> Median 312.236 -9.264 388.515 -9.429 365.292 39.930
#> Min - Max 187.77 - 501.87 -278.91 - 372.71 204.55 - 514.43 -190.58 - 173.87 200.19 - 493.40 -279.46 - 265.56
#> WEEK 3 DAY 22
#> n 15 15 15 15 15 15
#> Mean (SD) 342.062 (92.568) 6.768 (151.505) 326.684 (116.421) -36.420 (145.415) 366.245 (99.106) 18.935 (168.417)
#> Median 352.930 -22.771 298.353 -78.409 329.688 -21.584
#> Min - Max 199.40 - 476.04 -230.25 - 303.00 151.05 - 561.23 -205.30 - 293.76 249.42 - 580.81 -252.73 - 410.01
#> WEEK 4 DAY 29
#> n 15 15 15 15 15 15
#> Mean (SD) 371.650 (44.805) 36.356 (139.308) 333.697 (110.377) -29.407 (125.592) 333.181 (96.466) -14.130 (107.622)
#> Median 375.412 58.958 308.020 -40.987 330.911 -25.820
#> Min - Max 302.32 - 451.62 -214.07 - 258.04 183.09 - 531.08 -241.72 - 134.12 126.95 - 488.57 -234.92 - 152.49
#> WEEK 5 DAY 36
#> n 15 15 15 15 15 15
#> Mean (SD) 345.504 (130.543) 10.210 (198.224) 309.919 (84.624) -53.185 (105.730) 322.931 (67.801) -24.380 (117.331)
#> Median 355.730 -23.213 306.219 -12.373 341.988 -26.952
#> Min - Max 88.38 - 661.12 -271.06 - 539.84 189.01 - 448.58 -256.52 - 91.57 217.51 - 427.16 -291.03 - 171.19
#> RR Duration
#> BASELINE
#> n 15 15 15
#> Mean (SD) 1086.908 (363.811) 1050.034 (390.444) 1102.659 (310.359)
#> Median 1116.849 1089.193 1250.037
#> Min - Max 626.19 - 1653.12 414.61 - 1721.89 385.51 - 1430.81
#> WEEK 1 DAY 8
#> n 15 15 15 15 15 15
#> Mean (SD) 968.499 (287.811) -118.409 (546.796) 1041.186 (211.201) -8.848 (435.281) 948.491 (213.746) -154.168 (442.882)
#> Median 961.296 -147.460 1013.786 24.754 965.429 -224.054
#> Min - Max 358.92 - 1593.51 -1014.82 - 911.82 714.44 - 1417.52 -618.80 - 847.31 513.35 - 1229.09 -736.69 - 843.58
#> WEEK 2 DAY 15
#> n 15 15 15 15 15 15
#> Mean (SD) 932.717 (259.634) -154.191 (331.884) 1139.332 (454.231) 89.298 (582.750) 1021.283 (233.529) -81.376 (415.781)
#> Median 950.533 -205.949 1068.007 -5.449 964.616 -142.180
#> Min - Max 409.68 - 1269.35 -649.69 - 473.09 486.51 - 2048.73 -846.72 - 1148.61 667.36 - 1367.25 -647.47 - 616.15
#> WEEK 3 DAY 22
#> n 15 15 15 15 15 15
#> Mean (SD) 1068.865 (319.540) -18.043 (513.412) 1110.882 (259.523) 60.848 (432.700) 1105.918 (306.185) 3.259 (516.734)
#> Median 1201.998 -65.085 1163.690 51.200 1187.130 30.318
#> Min - Max 380.49 - 1551.65 -832.86 - 703.74 621.41 - 1453.29 -887.06 - 822.18 446.02 - 1648.32 -984.79 - 816.30
#> WEEK 4 DAY 29
#> n 15 15 15 15 15 15
#> Mean (SD) 1087.915 (205.940) 1.008 (403.039) 1161.681 (293.257) 111.647 (460.979) 992.134 (283.177) -110.525 (334.932)
#> Median 1084.658 146.611 1055.223 191.008 1028.997 -112.599
#> Min - Max 697.59 - 1499.17 -801.16 - 402.97 722.35 - 1762.04 -528.27 - 1191.83 497.14 - 1382.12 -597.95 - 757.99
#> WEEK 5 DAY 36
#> n 15 15 15 15 15 15
#> Mean (SD) 1016.880 (424.428) -70.027 (505.078) 1135.131 (224.684) 85.097 (497.679) 1089.527 (238.909) -13.132 (362.606)
#> Median 962.584 -142.925 1158.815 -9.553 1081.015 16.706
#> Min - Max 352.97 - 1843.86 -894.83 - 1162.79 714.34 - 1436.68 -843.41 - 992.34 699.72 - 1611.38 -696.03 - 561.53
ECG Abnormalities (Regardless of Abnormality at Baseline)
(EGT02_1)
1. ECG Abnormalities (Regardless of Abnormality at Baseline)
The egt02_1 template produces the
standard ECG abnormalities summary where the abnormalities are
summarized regardless of the abnormality at baseline.
run(egt02_1, syn_data)
#> Assessment A: Drug X B: Placebo C: Combination
#> Abnormality (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————
#> Heart Rate
#> Low 4/15 (26.7%) 4/15 (26.7%) 4/15 (26.7%)
#> High 4/15 (26.7%) 3/15 (20%) 3/15 (20%)
#> QT Duration
#> Low 2/15 (13.3%) 5/15 (33.3%) 3/15 (20%)
#> High 3/15 (20%) 6/15 (40%) 2/15 (13.3%)
#> RR Duration
#> Low 6/15 (40%) 2/15 (13.3%) 4/15 (26.7%)
#> High 4/15 (26.7%) 5/15 (33.3%) 2/15 (13.3%)
ECG Abnormalities (Among Subject Without Abnormality at
Baseline) (EGT02_2)
1. ECG Abnormalities (Among Subject Without Abnormality at Baseline)
The egt02_2 template produces the
standard ECG abnormalities summary where the abnormalities are
summarized among subject without abnormality at baseline.
run(egt02_2, syn_data)
#> Assessment A: Drug X B: Placebo C: Combination
#> Abnormality (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————
#> Heart Rate
#> Low 4/15 (26.7%) 4/14 (28.6%) 4/15 (26.7%)
#> High 3/13 (23.1%) 3/15 (20%) 2/14 (14.3%)
#> QT Duration
#> Low 2/12 (16.7%) 5/15 (33.3%) 3/14 (21.4%)
#> High 3/14 (21.4%) 6/15 (40%) 2/14 (14.3%)
#> RR Duration
#> Low 6/15 (40%) 2/13 (15.4%) 4/14 (28.6%)
#> High 4/13 (30.8%) 5/13 (38.5%) 2/15 (13.3%)
Shift Table of ECG Interval Data - Baseline versus
Minimum/Maximum Post-Baseline (EGT03)
1. Shift Table of ECG Interval Data - Baseline versus Minimum Post-Baseline
The egt03 template produces the
standard shift table of ECG interval data - baseline versus minimum
post-baseline summary.
proc_data <- log_filter(syn_data, PARAMCD == "HR", "adeg")
run(egt03, proc_data)
#> Actual Arm Code Minimum Post-Baseline Assessment
#> Baseline Reference Range Indicator LOW NORMAL HIGH Missing
#> ————————————————————————————————————————————————————————————————————————————————
#> Heart Rate
#> ARM A (N=15)
#> LOW 0 0 0 0
#> NORMAL 4 (26.7%) 9 (60.0%) 0 0
#> HIGH 0 2 (13.3%) 0 0
#> Missing 0 0 0 0
#> ARM B (N=15)
#> LOW 0 1 (6.7%) 0 0
#> NORMAL 4 (26.7%) 10 (66.7%) 0 0
#> HIGH 0 0 0 0
#> Missing 0 0 0 0
#> ARM C (N=15)
#> LOW 0 0 0 0
#> NORMAL 4 (26.7%) 10 (66.7%) 0 0
#> HIGH 0 1 (6.7%) 0 0
#> Missing 0 0 0 0
ECG Actual Values and Changes from Baseline by Visit
(EGT05_QTCAT)
1. ECG Actual Values and Changes from Baseline by Visit
The egt05_qtcat template produces the
standard ECG actual values and changes from baseline by visit
summary.
run(egt05_qtcat, syn_data)
#> Parameter
#> Analysis Visit A: Drug X B: Placebo C: Combination
#> Category (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————
#> QT Duration
#> BASELINE
#> Value at Visit
#> n 15 15 15
#> <=450 msec 13 (86.7%) 15 (100%) 13 (86.7%)
#> >450 to <=480 msec 1 (6.7%) 0 0
#> >480 to <=500 msec 0 0 1 (6.7%)
#> >500 msec 1 (6.7%) 0 1 (6.7%)
#> WEEK 1 DAY 8
#> Value at Visit
#> n 15 15 15
#> <=450 msec 12 (80.0%) 9 (60.0%) 13 (86.7%)
#> >450 to <=480 msec 1 (6.7%) 1 (6.7%) 1 (6.7%)
#> >480 to <=500 msec 1 (6.7%) 3 (20.0%) 1 (6.7%)
#> >500 msec 1 (6.7%) 2 (13.3%) 0
#> Change from Baseline
#> n 15 15 15
#> <=30 msec 7 (46.7%) 6 (40.0%) 9 (60.0%)
#> >30 to <=60 msec 2 (13.3%) 1 (6.7%) 1 (6.7%)
#> >60 msec 6 (40.0%) 8 (53.3%) 5 (33.3%)
#> WEEK 2 DAY 15
#> Value at Visit
#> n 15 15 15
#> <=450 msec 11 (73.3%) 14 (93.3%) 12 (80.0%)
#> >450 to <=480 msec 2 (13.3%) 0 2 (13.3%)
#> >480 to <=500 msec 1 (6.7%) 0 1 (6.7%)
#> >500 msec 1 (6.7%) 1 (6.7%) 0
#> Change from Baseline
#> n 15 15 15
#> <=30 msec 9 (60.0%) 12 (80.0%) 7 (46.7%)
#> >30 to <=60 msec 2 (13.3%) 0 3 (20.0%)
#> >60 msec 4 (26.7%) 3 (20.0%) 5 (33.3%)
#> WEEK 3 DAY 22
#> Value at Visit
#> n 15 15 15
#> <=450 msec 12 (80.0%) 12 (80.0%) 12 (80.0%)
#> >450 to <=480 msec 3 (20.0%) 1 (6.7%) 1 (6.7%)
#> >500 msec 0 2 (13.3%) 2 (13.3%)
#> Change from Baseline
#> n 15 15 15
#> <=30 msec 9 (60.0%) 11 (73.3%) 9 (60.0%)
#> >30 to <=60 msec 1 (6.7%) 1 (6.7%) 0
#> >60 msec 5 (33.3%) 3 (20.0%) 6 (40.0%)
#> WEEK 4 DAY 29
#> Value at Visit
#> n 15 15 15
#> <=450 msec 14 (93.3%) 12 (80.0%) 13 (86.7%)
#> >450 to <=480 msec 1 (6.7%) 1 (6.7%) 1 (6.7%)
#> >480 to <=500 msec 0 0 1 (6.7%)
#> >500 msec 0 2 (13.3%) 0
#> Change from Baseline
#> n 15 15 15
#> <=30 msec 6 (40.0%) 9 (60.0%) 9 (60.0%)
#> >30 to <=60 msec 2 (13.3%) 1 (6.7%) 2 (13.3%)
#> >60 msec 7 (46.7%) 5 (33.3%) 4 (26.7%)
#> WEEK 5 DAY 36
#> Value at Visit
#> n 15 15 15
#> <=450 msec 12 (80.0%) 15 (100%) 15 (100%)
#> >450 to <=480 msec 2 (13.3%) 0 0
#> >500 msec 1 (6.7%) 0 0
#> Change from Baseline
#> n 15 15 15
#> <=30 msec 9 (60.0%) 11 (73.3%) 9 (60.0%)
#> >30 to <=60 msec 0 3 (20.0%) 2 (13.3%)
#> >60 msec 6 (40.0%) 1 (6.7%) 4 (26.7%)2. ECG Actual Values and Changes from Baseline by Visit (removing default analyses)
The template have two default analyses of ADEG.AVALCAT1
and ADEG.CHGCAT1. To keep only the analyses needed, this
can be achieved by modifying the parameter summaryvars.
run(egt05_qtcat, syn_data, summaryvars = c("AVALCAT1"))
#> Parameter
#> Analysis Visit A: Drug X B: Placebo C: Combination
#> Category (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————
#> QT Duration
#> BASELINE
#> n 15 15 15
#> <=450 msec 13 (86.7%) 15 (100%) 13 (86.7%)
#> >450 to <=480 msec 1 (6.7%) 0 0
#> >480 to <=500 msec 0 0 1 (6.7%)
#> >500 msec 1 (6.7%) 0 1 (6.7%)
#> WEEK 1 DAY 8
#> n 15 15 15
#> <=450 msec 12 (80.0%) 9 (60.0%) 13 (86.7%)
#> >450 to <=480 msec 1 (6.7%) 1 (6.7%) 1 (6.7%)
#> >480 to <=500 msec 1 (6.7%) 3 (20.0%) 1 (6.7%)
#> >500 msec 1 (6.7%) 2 (13.3%) 0
#> WEEK 2 DAY 15
#> n 15 15 15
#> <=450 msec 11 (73.3%) 14 (93.3%) 12 (80.0%)
#> >450 to <=480 msec 2 (13.3%) 0 2 (13.3%)
#> >480 to <=500 msec 1 (6.7%) 0 1 (6.7%)
#> >500 msec 1 (6.7%) 1 (6.7%) 0
#> WEEK 3 DAY 22
#> n 15 15 15
#> <=450 msec 12 (80.0%) 12 (80.0%) 12 (80.0%)
#> >450 to <=480 msec 3 (20.0%) 1 (6.7%) 1 (6.7%)
#> >500 msec 0 2 (13.3%) 2 (13.3%)
#> WEEK 4 DAY 29
#> n 15 15 15
#> <=450 msec 14 (93.3%) 12 (80.0%) 13 (86.7%)
#> >450 to <=480 msec 1 (6.7%) 1 (6.7%) 1 (6.7%)
#> >480 to <=500 msec 0 0 1 (6.7%)
#> >500 msec 0 2 (13.3%) 0
#> WEEK 5 DAY 36
#> n 15 15 15
#> <=450 msec 12 (80.0%) 15 (100%) 15 (100%)
#> >450 to <=480 msec 2 (13.3%) 0 0
#> >500 msec 1 (6.7%) 0 0
Study Drug Exposure (EXT01)
1. Study Drug Exposure
- The
ext01template displays total number of doses administered and total dose administered by default - The template does not include the column of total by default
run(ext01, syn_data)
#> A: Drug X B: Placebo C: Combination
#> PARCAT2 (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————
#> Drug A
#> Overall duration (days)
#> n 11 7 7
#> Mean (SD) 157.5 (67.4) 115.4 (62.8) 98.6 (68.8)
#> Median 174.0 119.0 89.0
#> Min - Max 53.0 - 239.0 22.0 - 219.0 1.0 - 182.0
#> Total dose administered
#> n 11 7 7
#> Mean (SD) 6567.3 (1127.1) 7028.6 (1626.1) 6377.1 (863.7)
#> Median 6720.0 7200.0 6480.0
#> Min - Max 4800.0 - 8400.0 5280.0 - 9360.0 5280.0 - 7440.0
#> Drug B
#> Overall duration (days)
#> n 4 8 8
#> Mean (SD) 142.2 (100.3) 105.9 (60.0) 158.2 (96.2)
#> Median 160.0 95.0 203.0
#> Min - Max 17.0 - 232.0 37.0 - 211.0 27.0 - 249.0
#> Total dose administered
#> n 4 8 8
#> Mean (SD) 7020.0 (1148.9) 5250.0 (864.7) 5940.0 (1187.9)
#> Median 6960.0 5160.0 5880.0
#> Min - Max 5760.0 - 8400.0 4080.0 - 6480.0 4320.0 - 7680.0
Laboratory Test Results and Change from Baseline by Visit
(LBT01)
1. Laboratory Test Results and Change from Baseline by Visit
- The
lbt01template produces the standard laboratory test results and change from baseline by visit. - To select the SI/CV/LS results and the panel (chemistry/hematology/urinalysis/coagulation etc.) to display, user defines individual filters and apply to input datasets prior to running CHEVRON.
t_lb_chg <- run(lbt01, syn_data)
head(t_lb_chg, 20)
#> A: Drug X B: Placebo C: Combination
#> Change from Change from Change from
#> Value at Visit Baseline Value at Visit Baseline Value at Visit Baseline
#> (N=15) (N=15) (N=15) (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Alanine Aminotransferase Measurement
#> BASELINE
#> n 15 15 15
#> Mean (SD) 18.655 (12.455) 16.835 (11.080) 22.385 (9.452)
#> Median 16.040 17.453 25.250
#> Min - Max 2.43 - 44.06 1.48 - 31.99 0.57 - 37.23
#> WEEK 1 DAY 8
#> n 15 15 15 15 15 15
#> Mean (SD) 16.308 (10.850) -2.348 (17.558) 22.055 (7.537) 5.220 (16.359) 19.574 (9.876) -2.811 (10.902)
#> Median 14.664 -5.369 22.476 7.252 19.425 -0.995
#> Min - Max 0.10 - 36.30 -30.18 - 22.66 9.72 - 33.81 -16.82 - 32.33 1.03 - 36.28 -19.61 - 18.45
#> WEEK 2 DAY 15
#> n 15 15 15 15 15 15
#> Mean (SD) 16.646 (10.528) -2.010 (15.773) 20.758 (9.578) 3.923 (14.084) 10.911 (7.721) -11.474 (11.002)
#> Median 15.470 -6.427 18.499 6.248 9.850 -8.657
#> Min - Max 0.40 - 35.29 -29.99 - 32.86 1.56 - 42.84 -24.92 - 29.85 0.35 - 25.01 -27.38 - 2.52
#> WEEK 3 DAY 22
#> n 15 15 15 15 15 15
#> Mean (SD) 17.488 (10.679) -1.167 (15.759) 20.055 (8.086) 3.219 (16.285) 18.413 (9.513) -3.973 (9.966)
#> Median 14.224 1.355 21.852 5.345 19.529 -7.194
Laboratory Abnormalities (LBT04)
1. Laboratory Abnormalities
- The
lbt04template produces the standard laboratory abnormalities summary. - The template subsets to SI results by default.
- The laboratory tests and directions of abnormality in this template is data-driven. Table entries provide the number of patients with a during treatment laboratory value abnormality in the direction specified among patients without this abnormality at baseline.
run(lbt04, syn_data)
#> Laboratory Test A: Drug X B: Placebo C: Combination
#> Direction of Abnormality (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————
#> CHEMISTRY
#> Alanine Aminotransferase Measurement
#> Low 0/7 0/2 1/7 (14.3%)
#> High 0/7 0/3 0/8
#> C-Reactive Protein Measurement
#> Low 0/8 1/2 (50.0%) 0/6
#> High 3/8 (37.5%) 0/2 0/7
#> Immunoglobulin A Measurement
#> Low 0/5 0/8 0/7
#> High 1/3 (33.3%) 1/8 (12.5%) 0/6
#> COAGULATION
#> Alanine Aminotransferase Measurement
#> Low 0/3 0/6 0/4
#> High 0/5 0/7 0/4
#> C-Reactive Protein Measurement
#> Low 0/5 0/5 1/3 (33.3%)
#> High 0/5 1/6 (16.7%) 1/4 (25.0%)
#> Immunoglobulin A Measurement
#> Low 0/8 0/9 0/6
#> High 0/8 0/9 1/6 (16.7%)
#> HEMATOLOGY
#> Alanine Aminotransferase Measurement
#> Low 0/4 0/5 0/4
#> High 0/6 0/5 0/4
#> C-Reactive Protein Measurement
#> Low 0/5 0/4 0/3
#> High 0/5 0/4 0/5
#> Immunoglobulin A Measurement
#> Low 0/3 0/4 0/8
#> High 0/3 0/4 0/7
Laboratory Abnormalities with Single and Replicated Marked
(LBT05)
1. Laboratory Abnormalities with Single and Replicated Marked
- The
lbt05template produces the standard laboratory abnormalities summary for marked abnormalities. - The laboratory tests and directions of abnormality in this template is currently data-driven. The standard metadata for Safety Lab Standardization will be incorporated in future release.
run(lbt05, syn_data)
#> Laboratory Test A: Drug X B: Placebo C: Combination
#> Direction of Abnormality (N=15) (N=15) (N=15)
#> ——————————————————————————————————————————————————————————————————————————————————
#> Alanine Aminotransferase Measurement (n) 15 14 14
#> Low
#> Single, not last 1 (6.7%) 0 4 (28.6%)
#> Last or replicated 5 (33.3%) 4 (28.6%) 4 (28.6%)
#> Any Abnormality 6 (40.0%) 4 (28.6%) 8 (57.1%)
#> High
#> Single, not last 0 0 0
#> Last or replicated 0 0 0
#> Any Abnormality 0 0 0
#> C-Reactive Protein Measurement (n) 15 15 15
#> Low
#> Single, not last 4 (26.7%) 0 3 (20.0%)
#> Last or replicated 3 (20.0%) 5 (33.3%) 6 (40.0%)
#> Any Abnormality 7 (46.7%) 5 (33.3%) 9 (60.0%)
#> High
#> Single, not last 1 (6.7%) 3 (20.0%) 0
#> Last or replicated 4 (26.7%) 3 (20.0%) 6 (40.0%)
#> Any Abnormality 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> Immunoglobulin A Measurement (n) 13 14 14
#> Low
#> Single, not last 0 0 0
#> Last or replicated 0 0 0
#> Any Abnormality 0 0 0
#> High
#> Single, not last 6 (46.2%) 1 (7.1%) 2 (14.3%)
#> Last or replicated 3 (23.1%) 4 (28.6%) 3 (21.4%)
#> Any Abnormality 9 (69.2%) 5 (35.7%) 5 (35.7%)
Laboratory Abnormalities by Visit and Baseline Status
(LBT06)
1. Laboratory Abnormalities by Visit and Baseline Status
- The
lbt06template produces the standard laboratory abnormalities by visit and baseline status summary.
run(lbt06, syn_data)
#> Visit
#> Abnormality at Visit A: Drug X B: Placebo C: Combination
#> Baseline Status (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————
#> Alanine Aminotransferase Measurement
#> WEEK 1 DAY 8
#> Low
#> Not low 0/1 0/6 0/1
#> Low 0/1 0/1 0/1
#> Total 0/2 0/7 0/2
#> High
#> Not high 0/2 0/7 0/2
#> High 0/0 0/0 0/0
#> Total 0/2 0/7 0/2
#> WEEK 2 DAY 15
#> Low
#> Not low 0/3 0/2 0/2
#> Low 0/0 0/0 0/0
#> Total 0/3 0/2 0/2
#> High
#> Not high 0/3 0/2 0/2
#> High 0/0 0/0 0/0
#> Total 0/3 0/2 0/2
#> WEEK 3 DAY 22
#> Low
#> Not low 0/5 0/3 1/6 (16.7%)
#> Low 0/0 0/0 0/0
#> Total 0/5 0/3 1/6 (16.7%)
#> High
#> Not high 0/5 0/3 0/6
#> High 0/0 0/0 0/0
#> Total 0/5 0/3 0/6
#> WEEK 4 DAY 29
#> Low
#> Not low 0/3 0/1 0/1
#> Low 0/3 0/2 0/0
#> Total 0/6 0/3 0/1
#> High
#> Not high 0/6 0/3 0/1
#> High 0/0 0/0 0/0
#> Total 0/6 0/3 0/1
#> WEEK 5 DAY 36
#> Low
#> Not low 0/2 0/2 0/5
#> Low 0/1 0/1 0/0
#> Total 0/3 0/3 0/5
#> High
#> Not high 0/3 0/3 0/5
#> High 0/0 0/0 0/0
#> Total 0/3 0/3 0/5
#> C-Reactive Protein Measurement
#> WEEK 1 DAY 8
#> Low
#> Not low 0/5 0/3 0/3
#> Low 0/0 0/1 0/0
#> Total 0/5 0/4 0/3
#> High
#> Not high 0/5 0/3 1/3 (33.3%)
#> High 0/0 0/1 0/0
#> Total 0/5 0/4 1/3 (33.3%)
#> WEEK 2 DAY 15
#> Low
#> Not low 0/8 0/2 0/0
#> Low 0/0 0/0 0/1
#> Total 0/8 0/2 0/1
#> High
#> Not high 1/8 (12.5%) 0/1 0/1
#> High 0/0 0/1 0/0
#> Total 1/8 (12.5%) 0/2 0/1
#> WEEK 3 DAY 22
#> Low
#> Not low 0/5 0/4 0/4
#> Low 0/0 1/1 (100%) 0/2
#> Total 0/5 1/5 (20%) 0/6
#> High
#> Not high 1/5 (20%) 1/5 (20%) 0/6
#> High 0/0 0/0 0/0
#> Total 1/5 (20%) 1/5 (20%) 0/6
#> WEEK 4 DAY 29
#> Low
#> Not low 0/2 1/2 (50%) 1/3 (33.3%)
#> Low 0/0 0/0 0/0
#> Total 0/2 1/2 (50%) 1/3 (33.3%)
#> High
#> Not high 0/2 0/2 0/3
#> High 0/0 0/0 0/0
#> Total 0/2 0/2 0/3
#> WEEK 5 DAY 36
#> Low
#> Not low 0/2 0/2 0/5
#> Low 0/0 1/1 (100%) 0/1
#> Total 0/2 1/3 (33.3%) 0/6
#> High
#> Not high 1/2 (50%) 0/3 0/6
#> High 0/0 0/0 0/0
#> Total 1/2 (50%) 0/3 0/6
#> Immunoglobulin A Measurement
#> WEEK 1 DAY 8
#> Low
#> Not low 0/6 0/6 0/2
#> Low 0/0 0/0 0/0
#> Total 0/6 0/6 0/2
#> High
#> Not high 0/5 1/6 (16.7%) 0/2
#> High 0/1 0/0 0/0
#> Total 0/6 1/6 (16.7%) 0/2
#> WEEK 2 DAY 15
#> Low
#> Not low 0/3 0/7 0/4
#> Low 0/0 0/0 0/0
#> Total 0/3 0/7 0/4
#> High
#> Not high 0/3 0/7 1/4 (25%)
#> High 0/0 0/0 0/0
#> Total 0/3 0/7 1/4 (25%)
#> WEEK 3 DAY 22
#> Low
#> Not low 0/4 0/5 0/9
#> Low 0/0 0/0 0/0
#> Total 0/4 0/5 0/9
#> High
#> Not high 0/3 0/5 0/8
#> High 0/1 0/0 0/1
#> Total 0/4 0/5 0/9
#> WEEK 4 DAY 29
#> Low
#> Not low 0/2 0/6 0/4
#> Low 0/0 0/0 0/0
#> Total 0/2 0/6 0/4
#> High
#> Not high 1/1 (100%) 0/6 0/3
#> High 0/1 0/0 0/1
#> Total 1/2 (50%) 0/6 0/4
#> WEEK 5 DAY 36
#> Low
#> Not low 0/6 0/5 0/5
#> Low 0/0 0/0 0/0
#> Total 0/6 0/5 0/5
#> High
#> Not high 0/5 0/5 0/5
#> High 0/1 0/0 0/0
#> Total 0/6 0/5 0/5
Laboratory Test Results with Highest NCI CTCAE
Grade Post-Baseline (LBT07)
1. Laboratory Test Results with Highest
NCI CTCAE Grade Post-Baseline
- The
lbt07template produces the standard laboratory test results with highestNCI CTCAEgrade post-baseline summary. - The laboratory tests and grades in this template is currently
data-driven. The standard metadata for possible lab tests and
corresponding
NCI CTCAEgrade will be incorporated in future release.
run(lbt07, syn_data)
#> Parameter
#> Direction of Abnormality A: Drug X B: Placebo C: Combination
#> Highest NCI CTCAE Grade (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————
#> Alanine Aminotransferase Measurement (n) 15 15 15
#> LOW
#> 1 3 (20.0%) 0 0
#> 2 2 (13.3%) 1 (6.7%) 1 (6.7%)
#> 3 1 (6.7%) 1 (6.7%) 6 (40.0%)
#> 4 3 (20.0%) 2 (13.3%) 3 (20.0%)
#> Any 9 (60.0%) 4 (26.7%) 10 (66.7%)
#> C-Reactive Protein Measurement (n) 15 15 15
#> LOW
#> 1 2 (13.3%) 1 (6.7%) 2 (13.3%)
#> 2 5 (33.3%) 2 (13.3%) 5 (33.3%)
#> 3 3 (20.0%) 4 (26.7%) 3 (20.0%)
#> 4 0 1 (6.7%) 0
#> Any 10 (66.7%) 8 (53.3%) 10 (66.7%)
#> HIGH
#> 1 3 (20.0%) 1 (6.7%) 1 (6.7%)
#> 2 4 (26.7%) 4 (26.7%) 2 (13.3%)
#> 3 1 (6.7%) 2 (13.3%) 4 (26.7%)
#> 4 0 1 (6.7%) 0
#> Any 8 (53.3%) 8 (53.3%) 7 (46.7%)
#> Immunoglobulin A Measurement (n) 15 15 15
#> HIGH
#> 1 3 (20.0%) 1 (6.7%) 1 (6.7%)
#> 2 5 (33.3%) 4 (26.7%) 2 (13.3%)
#> 3 3 (20.0%) 3 (20.0%) 2 (13.3%)
#> 4 0 0 1 (6.7%)
#> Any 11 (73.3%) 8 (53.3%) 6 (40.0%)
Laboratory Test Results Shift Table - Highest
NCI-CTCAE Grade Post-Baseline by Baseline
NCI-CTCAE Grade (LBT14)
1. Laboratory Test Results Shift Table - Highest
NCI-CTCAE Grade Post-Baseline by Baseline
NCI-CTCAE Grade (High)
To produce the standard laboratory test results shift table - highest
NCI-CTCAE grade post-baseline by baseline
NCI-CTCAE grade summary for high abnormalities, use the
lbt14 template and set the parameter
direction to high.
run(lbt14, syn_data, direction = "high")
#> Baseline Toxicity Grade A: Drug X B: Placebo C: Combination
#> Post-baseline NCI-CTCAE Grade (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————
#> Alanine Aminotransferase Measurement
#> Not High 15 15 15
#> Not High 15 (100%) 15 (100%) 15 (100%)
#> C-Reactive Protein Measurement
#> Not High 15 13 14
#> Not High 7 (46.7%) 7 (53.8%) 8 (57.1%)
#> 1 3 (20.0%) 1 (7.7%) 1 (7.1%)
#> 2 4 (26.7%) 3 (23.1%) 1 (7.1%)
#> 3 1 (6.7%) 1 (7.7%) 4 (28.6%)
#> 4 0 1 (7.7%) 0
#> 1 0 0 1
#> 2 0 0 1 (100%)
#> 3 0 1 0
#> 2 0 1 (100%) 0
#> 4 0 1 0
#> 3 0 1 (100%) 0
#> Immunoglobulin A Measurement
#> Not High 12 14 13
#> Not High 3 (25.0%) 7 (50.0%) 8 (61.5%)
#> 1 3 (25.0%) 1 (7.1%) 1 (7.7%)
#> 2 3 (25.0%) 3 (21.4%) 2 (15.4%)
#> 3 3 (25.0%) 3 (21.4%) 2 (15.4%)
#> 1 2 0 1
#> Not High 1 (50.0%) 0 1 (100%)
#> 2 1 (50.0%) 0 0
#> 3 0 0 1
#> 4 0 0 1 (100%)
#> 4 1 1 0
#> 2 1 (100%) 1 (100%) 0
2. Laboratory Test Results Shift Table - Highest
NCI-CTCAE Grade Post-Baseline by Baseline
NCI-CTCAE Grade (Low)
To produce the standard laboratory test results shift table - highest
NCI-CTCAE grade post-baseline by baseline
NCI-CTCAE grade summary for high abnormalities, use the
lbt14 template and the argument
direction is low by default.
run(lbt14, syn_data)
#> Baseline Toxicity Grade A: Drug X B: Placebo C: Combination
#> Post-baseline NCI-CTCAE Grade (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————
#> Alanine Aminotransferase Measurement
#> Not Low 12 12 14
#> Not Low 5 (41.7%) 8 (66.7%) 5 (35.7%)
#> 1 3 (25.0%) 0 0
#> 2 2 (16.7%) 1 (8.3%) 1 (7.1%)
#> 3 0 1 (8.3%) 5 (35.7%)
#> 4 2 (16.7%) 2 (16.7%) 3 (21.4%)
#> 1 1 2 0
#> Not Low 1 (100%) 2 (100%) 0
#> 2 1 1 0
#> Not Low 0 1 (100%) 0
#> 4 1 (100%) 0 0
#> 3 1 0 1
#> 3 1 (100%) 0 1 (100%)
#> C-Reactive Protein Measurement
#> Not Low 14 13 12
#> Not Low 5 (35.7%) 7 (53.8%) 4 (33.3%)
#> 1 2 (14.3%) 0 2 (16.7%)
#> 2 5 (35.7%) 2 (15.4%) 4 (33.3%)
#> 3 2 (14.3%) 3 (23.1%) 2 (16.7%)
#> 4 0 1 (7.7%) 0
#> 1 0 0 2
#> Not Low 0 0 1 (50.0%)
#> 2 0 0 1 (50.0%)
#> 2 0 1 0
#> 1 0 1 (100%) 0
#> 3 1 1 1
#> 3 1 (100%) 1 (100%) 1 (100%)
#> Immunoglobulin A Measurement
#> Not Low 15 15 15
#> Not Low 15 (100%) 15 (100%) 15 (100%)
3. Laboratory Test Results Shift Table - Highest
NCI-CTCAE Grade Post-Baseline by Baseline
NCI-CTCAE Grade (High) Without Patients with Missing
Baseline
To exclude patients with missing baseline grade, set the argument
gr_missing to excl.
run(lbt14, syn_data, direction = "high", gr_missing = "excl")
#> Baseline Toxicity Grade A: Drug X B: Placebo C: Combination
#> Post-baseline NCI-CTCAE Grade (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————
#> Alanine Aminotransferase Measurement
#> Not High 15 15 15
#> Not High 15 (100%) 15 (100%) 15 (100%)
#> C-Reactive Protein Measurement
#> Not High 15 13 14
#> Not High 7 (46.7%) 7 (53.8%) 8 (57.1%)
#> 1 3 (20.0%) 1 (7.7%) 1 (7.1%)
#> 2 4 (26.7%) 3 (23.1%) 1 (7.1%)
#> 3 1 (6.7%) 1 (7.7%) 4 (28.6%)
#> 4 0 1 (7.7%) 0
#> 1 0 0 1
#> 2 0 0 1 (100%)
#> 3 0 1 0
#> 2 0 1 (100%) 0
#> 4 0 1 0
#> 3 0 1 (100%) 0
#> Immunoglobulin A Measurement
#> Not High 12 14 13
#> Not High 3 (25.0%) 7 (50.0%) 8 (61.5%)
#> 1 3 (25.0%) 1 (7.1%) 1 (7.7%)
#> 2 3 (25.0%) 3 (21.4%) 2 (15.4%)
#> 3 3 (25.0%) 3 (21.4%) 2 (15.4%)
#> 1 2 0 1
#> Not High 1 (50.0%) 0 1 (100%)
#> 2 1 (50.0%) 0 0
#> 3 0 0 1
#> 4 0 0 1 (100%)
#> 4 1 1 0
#> 2 1 (100%) 1 (100%) 0
4. Laboratory Test Results Shift Table - Highest
NCI-CTCAE Grade Post-Baseline by Baseline
NCI-CTCAE Grade (Low) with Missing Baseline Considered as
Grade 0
To count patients with missing baseline grade as grade 0, set the
argument gr_missing to gr_0.
run(lbt14, syn_data, gr_missing = "gr_0")
#> Baseline Toxicity Grade A: Drug X B: Placebo C: Combination
#> Post-baseline NCI-CTCAE Grade (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————
#> Alanine Aminotransferase Measurement
#> 1 1 2 0
#> Not Low 1 (100%) 2 (100%) 0
#> 2 1 1 0
#> Not Low 0 1 (100%) 0
#> 4 1 (100%) 0 0
#> 3 1 0 1
#> 3 1 (100%) 0 1 (100%)
#> Not Low 12 12 14
#> Not Low 5 (41.7%) 8 (66.7%) 5 (35.7%)
#> 1 3 (25.0%) 0 0
#> 2 2 (16.7%) 1 (8.3%) 1 (7.1%)
#> 3 0 1 (8.3%) 5 (35.7%)
#> 4 2 (16.7%) 2 (16.7%) 3 (21.4%)
#> C-Reactive Protein Measurement
#> 1 0 0 2
#> 1 0 0 1 (50.0%)
#> 3 0 0 1 (50.0%)
#> 2 0 1 0
#> 2 0 1 (100%) 0
#> 3 1 1 1
#> 3 1 (100%) 1 (100%) 1 (100%)
#> Not Low 14 13 12
#> Not Low 5 (35.7%) 7 (53.8%) 4 (33.3%)
#> 1 2 (14.3%) 0 2 (16.7%)
#> 2 5 (35.7%) 2 (15.4%) 4 (33.3%)
#> 3 2 (14.3%) 3 (23.1%) 2 (16.7%)
#> 4 0 1 (7.7%) 0
#> Immunoglobulin A Measurement
#> Not Low 15 15 15
#> Not Low 15 (100%) 15 (100%) 15 (100%)
4. Laboratory Test Results Shift Table - Highest
NCI-CTCAE Grade Post-Baseline by Baseline
NCI-CTCAE Grade (with fill in of grades)
To display all possible grades even if they do not occur in the data,
set the argument prune_0 to FALSE.
run(lbt14, syn_data, direction = "high", prune_0 = FALSE)
#> Baseline Toxicity Grade A: Drug X B: Placebo C: Combination
#> Post-baseline NCI-CTCAE Grade (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————
#> Alanine Aminotransferase Measurement
#> Not High 15 15 15
#> Not High 15 (100%) 15 (100%) 15 (100%)
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> 1 0 0 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> 2 0 0 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> 3 0 0 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> 4 0 0 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> Missing 0 0 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> C-Reactive Protein Measurement
#> Not High 15 13 14
#> Not High 7 (46.7%) 7 (53.8%) 8 (57.1%)
#> 1 3 (20.0%) 1 (7.7%) 1 (7.1%)
#> 2 4 (26.7%) 3 (23.1%) 1 (7.1%)
#> 3 1 (6.7%) 1 (7.7%) 4 (28.6%)
#> 4 0 1 (7.7%) 0
#> Missing 0 0 0
#> 1 0 0 1
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 1 (100%)
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> 2 0 0 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> 3 0 1 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 1 (100%) 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> 4 0 1 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 1 (100%) 0
#> 4 0 0 0
#> Missing 0 0 0
#> Missing 0 0 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> Immunoglobulin A Measurement
#> Not High 12 14 13
#> Not High 3 (25.0%) 7 (50.0%) 8 (61.5%)
#> 1 3 (25.0%) 1 (7.1%) 1 (7.7%)
#> 2 3 (25.0%) 3 (21.4%) 2 (15.4%)
#> 3 3 (25.0%) 3 (21.4%) 2 (15.4%)
#> 4 0 0 0
#> Missing 0 0 0
#> 1 2 0 1
#> Not High 1 (50.0%) 0 1 (100%)
#> 1 0 0 0
#> 2 1 (50.0%) 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> 2 0 0 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> 3 0 0 1
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 1 (100%)
#> Missing 0 0 0
#> 4 1 1 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 1 (100%) 1 (100%) 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
#> Missing 0 0 0
#> Not High 0 0 0
#> 1 0 0 0
#> 2 0 0 0
#> 3 0 0 0
#> 4 0 0 0
#> Missing 0 0 0
Medical History (MHT01)
1. Medical History
- The
mht01template displays medical conditions by MedDRA system organ class and Preferred Name by default. - The default treatment variable is
"ADSL.ARM". - The user is expected to use filter to subset medical conditions prior to or on entering study.
- By default, the template produces the overall ‘total number of conditions’ as well as the ‘total number of conditions’ per body system after the summary of patients. 5)This template currently does not support sorting MedDRA system organ class and preferred names by order of frequency.
run(mht01, syn_data)
#> MedDRA System Organ Class A: Drug X B: Placebo C: Combination
#> MedDRA Preferred Term (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one condition 13 (86.7%) 14 (93.3%) 15 (100%)
#> Total number of conditions 58 59 99
#> cl A
#> Total number of patients with at least one condition 7 (46.7%) 6 (40.0%) 10 (66.7%)
#> Total number of conditions 8 11 16
#> trm A_2/2 5 (33.3%) 6 (40.0%) 6 (40.0%)
#> trm A_1/2 3 (20.0%) 1 (6.7%) 6 (40.0%)
#> cl B
#> Total number of patients with at least one condition 12 (80.0%) 11 (73.3%) 12 (80.0%)
#> Total number of conditions 24 21 32
#> trm B_3/3 8 (53.3%) 6 (40.0%) 7 (46.7%)
#> trm B_1/3 5 (33.3%) 6 (40.0%) 8 (53.3%)
#> trm B_2/3 5 (33.3%) 6 (40.0%) 5 (33.3%)
#> cl C
#> Total number of patients with at least one condition 8 (53.3%) 6 (40.0%) 11 (73.3%)
#> Total number of conditions 10 13 22
#> trm C_2/2 6 (40.0%) 4 (26.7%) 8 (53.3%)
#> trm C_1/2 4 (26.7%) 4 (26.7%) 5 (33.3%)
#> cl D
#> Total number of patients with at least one condition 10 (66.7%) 7 (46.7%) 13 (86.7%)
#> Total number of conditions 16 14 29
#> trm D_1/3 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> trm D_2/3 6 (40.0%) 2 (13.3%) 7 (46.7%)
#> trm D_3/3 2 (13.3%) 5 (33.3%) 7 (46.7%)2. Medical History showing additional column ‘All Patients’
run(mht01, syn_data, lbl_overall = "All Patients")
#> MedDRA System Organ Class A: Drug X B: Placebo C: Combination All Patients
#> MedDRA Preferred Term (N=15) (N=15) (N=15) (N=45)
#> ————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one condition 13 (86.7%) 14 (93.3%) 15 (100%) 42 (93.3%)
#> Total number of conditions 58 59 99 216
#> cl A
#> Total number of patients with at least one condition 7 (46.7%) 6 (40.0%) 10 (66.7%) 23 (51.1%)
#> Total number of conditions 8 11 16 35
#> trm A_2/2 5 (33.3%) 6 (40.0%) 6 (40.0%) 17 (37.8%)
#> trm A_1/2 3 (20.0%) 1 (6.7%) 6 (40.0%) 10 (22.2%)
#> cl B
#> Total number of patients with at least one condition 12 (80.0%) 11 (73.3%) 12 (80.0%) 35 (77.8%)
#> Total number of conditions 24 21 32 77
#> trm B_3/3 8 (53.3%) 6 (40.0%) 7 (46.7%) 21 (46.7%)
#> trm B_1/3 5 (33.3%) 6 (40.0%) 8 (53.3%) 19 (42.2%)
#> trm B_2/3 5 (33.3%) 6 (40.0%) 5 (33.3%) 16 (35.6%)
#> cl C
#> Total number of patients with at least one condition 8 (53.3%) 6 (40.0%) 11 (73.3%) 25 (55.6%)
#> Total number of conditions 10 13 22 45
#> trm C_2/2 6 (40.0%) 4 (26.7%) 8 (53.3%) 18 (40.0%)
#> trm C_1/2 4 (26.7%) 4 (26.7%) 5 (33.3%) 13 (28.9%)
#> cl D
#> Total number of patients with at least one condition 10 (66.7%) 7 (46.7%) 13 (86.7%) 30 (66.7%)
#> Total number of conditions 16 14 29 59
#> trm D_1/3 4 (26.7%) 4 (26.7%) 7 (46.7%) 15 (33.3%)
#> trm D_2/3 6 (40.0%) 2 (13.3%) 7 (46.7%) 15 (33.3%)
#> trm D_3/3 2 (13.3%) 5 (33.3%) 7 (46.7%) 14 (31.1%)
Major Protocol Deviations (PDT01)
1. Major Protocol Deviations
- The
pdt01template produces the standard major protocol deviations output. - Users are expected to filter
addvto only include records whereDVCAT == "MAJOR"in pre-processing.
proc_data <- syn_data
proc_data$addv <- proc_data$addv %>%
filter(DVCAT == "MAJOR")
run(pdt01, proc_data)
#> Category A: Drug X B: Placebo C: Combination
#> Description (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one major protocol deviation 2 (13.3%) 4 (26.7%) 0
#> Total number of major protocol deviations 2 5 0
#> EXCLUSION CRITERIA
#> Active or untreated or other excluded cns metastases 0 1 (6.7%) 0
#> Pregnancy criteria 0 1 (6.7%) 0
#> INCLUSION CRITERIA
#> Ineligible cancer type or current cancer stage 1 (6.7%) 0 0
#> MEDICATION
#> Discontinued study drug for unspecified reason 0 1 (6.7%) 0
#> Received prohibited concomitant medication 0 1 (6.7%) 0
#> PROCEDURAL
#> Eligibility-related test not done/out of window 0 1 (6.7%) 0
#> Failure to sign updated ICF within two visits 1 (6.7%) 0 0
Reasons for Major Protocol Deviations Related to
Epidemic/Pandemic (PDT02)
1. Reasons for Major Protocol Deviations Related to Epidemic/Pandemic
- The
pdt02template produces the reasons for major protocol deviations related to epidemic/pandemic summary. - By default,
ADDV.DVREASprovides the reason andADDV.DVTERMprovides the description. - By default,
addvhas been filtered to include only records that meet the conditionAEPRELFL == "Y" & DVCAT == "MAJOR".
run(pdt02, syn_data)
#> Primary Reason A: Drug X B: Placebo C: Combination
#> Description (N=15) (N=15) (N=15)
#> ——————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Total number of patients with at least one major protocol deviation related to epidemic/pandemic 1 (6.7%) 0 0
#> Total number of major protocol deviations related to epidemic/pandemic 1 0 0
#> Site action due to epidemic/pandemic 1 (6.7%) 0 0
#> Failure to sign updated ICF within two visits 1 (6.7%) 0 0
Duration of Exposure for Risk Management Plan
(RMPT01)
1. Duration of Exposure for Risk Management Plan
The rmpt01 template produces the
standard duration of exposure output for the Risk Management Plan
(RMP).
Person time is the sum of exposure across all patients in days.
run(rmpt01, syn_data)
#> Patients Person time
#> Duration of exposure (N=45) (N=45)
#> —————————————————————————————————————————————————————————————
#> < 1 month 4 (8.9%) 67
#> 1 to <3 months 13 (28.9%) 837
#> 3 to <6 months 13 (28.9%) 1728
#> >=6 months 15 (33.3%) 3281
#> Total patients number/person time 45 (100.0%) 5913
Extent of Exposure by Age Group and Gender for Risk
Management Plan (RMPT03)
1. Extent of Exposure by Age Group and Gender for Risk Management Plan
The rmpt03 template produces the
standard extent of exposure by age group and gender output for the Risk
Management Plan (RMP).
By default, the AGEGR1 variable is used as the age
group. If AGEGR1 is available in ADSL only but
not in ADEX, it needs to be added to ADEX
first.
proc_data <- syn_data
proc_data <- propagate(proc_data, "adsl", "AGEGR1", "USUBJID")
#>
#> Updating: adae with: AGEGR1
#> Updating: adsaftte with: AGEGR1
#> Updating: adcm with: AGEGR1
#> Updating: addv with: AGEGR1
#> Updating: adeg with: AGEGR1
#> Updating: adex with: AGEGR1
#> Updating: adlb with: AGEGR1
#> Updating: admh with: AGEGR1
#> Skipping: adrs
#> Updating: adsub with: AGEGR1
#> Skipping: adtte
#> Updating: advs with: AGEGR1
run(rmpt03, proc_data)
#> F M All Genders
#> Patients Person time Patients Person time Patients Person time
#> Age Group (N=30) (N=30) (N=15) (N=15) (N=45) (N=45)
#> —————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> <65 30 (100.0%) 4088 15 (100.0%) 1825 45 (100.0%) 5913
#> Total patients number/person time 30 (100.0%) 4088 15 (100.0%) 1825 45 (100.0%) 5913Any other study specific age group can be used by editing the
parameter summaryvars. For all RMP tables, if
the variable specified per summaryvars is unavailable in
ADEX, it needs to be added to ADEX first.
proc_data <- syn_data
proc_data$adsl <- proc_data$adsl %>%
mutate(
AGEGR2 = with_label(
factor(case_when(
AAGE < 18 ~ "<18",
AAGE >= 18 & AAGE <= 65 ~ "18 - 65",
AAGE > 65 ~ ">65",
), levels = c("<18", "18 - 65", ">65")),
"Age Group 2"
)
)
proc_data <- propagate(proc_data, "adsl", "AGEGR2", "USUBJID")
#>
#> Updating: adae with: AGEGR2
#> Updating: adsaftte with: AGEGR2
#> Updating: adcm with: AGEGR2
#> Updating: addv with: AGEGR2
#> Updating: adeg with: AGEGR2
#> Updating: adex with: AGEGR2
#> Updating: adlb with: AGEGR2
#> Updating: admh with: AGEGR2
#> Updating: adrs with: AGEGR2
#> Updating: adsub with: AGEGR2
#> Updating: adtte with: AGEGR2
#> Updating: advs with: AGEGR2
run(rmpt03, proc_data, summaryvars = "AGEGR2")
#> F M All Genders
#> Patients Person time Patients Person time Patients Person time
#> Age Group 2 (N=30) (N=30) (N=15) (N=15) (N=45) (N=45)
#> —————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> 18 - 65 30 (100.0%) 4088 15 (100.0%) 1825 45 (100.0%) 5913
#> Total patients number/person time 30 (100.0%) 4088 15 (100.0%) 1825 45 (100.0%) 5913
Extent of Exposure by Ethnic Origin for Risk Management Plan
(RMPT04)
1. Extent of Exposure by Ethnic Origin for Risk Management Plan
The rmpt04 template produces the
standard extent of exposure by ethnic origin output for the Risk
Management Plan (RMP).
run(rmpt04, syn_data)
#> Patients Person time
#> ETHNIC (N=45) (N=45)
#> —————————————————————————————————————————————————————————————
#> HISPANIC OR LATINO 2 (4.4%) 309
#> NOT HISPANIC OR LATINO 41 (91.1%) 5555
#> NOT REPORTED 2 (4.4%) 49
#> Total patients number/person time 45 (100.0%) 5913
Extent of Exposure by Race for Risk Management Plan
(RMPT05)
1. Extent of Exposure by Race for Risk Management Plan
The rmpt05 template produces the
standard extent of exposure by race output for the Risk Management Plan
(RMP).
run(rmpt05, syn_data)
#> Patients Person time
#> RACE (N=45) (N=45)
#> —————————————————————————————————————————————————————————————
#> ASIAN 26 (57.8%) 3309
#> BLACK OR AFRICAN AMERICAN 9 (20.0%) 1139
#> WHITE 7 (15.6%) 1231
#> AMERICAN INDIAN OR ALASKA NATIVE 3 (6.7%) 234
#> Total patients number/person time 45 (100.0%) 5913
Best Overall Response (RSPT01)
1. Best Overall Response
- The
rspt01template produces the standard best overall response output. - The template syntax is built based on
RECIST 1.1. By default, the subjects with response results of"CR"or"PR"are considered as responders. - Users are expected to pre-process the input analysis data and select
the parameter to be analyzed, i.e., best overall response by
investigator or best overall response by
BICR. - Unstratified analysis is provided by default.
proc_data <- log_filter(syn_data, PARAMCD == "BESRSPI", "adrs")
run(rspt01, proc_data, ref_group = NULL, perform_analysis = "unstrat", strata = NULL)
#> Warning in stats::prop.test(tbl, correct = FALSE): Chi-squared approximation
#> may be incorrect
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————————
#> Responders 10 (66.7%) 9 (60.0%) 11 (73.3%)
#> 95% CI (Wald, with correction) (39.5, 93.9) (31.9, 88.1) (47.6, 99.0)
#> Unstratified Analysis
#> Difference in Response rate (%) -6.7 6.7
#> 95% CI (Wald, with correction) (-47.7, 34.4) (-32.7, 46.0)
#> p-value (Chi-Squared Test) 0.7048 0.6903
#> Odds Ratio (95% CI) 0.75 (0.17 - 3.33) 1.37 (0.29 - 6.60)
#> Complete Response (CR) 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> 95% CI (Wald, with correction) (0.95, 52.38) (0.95, 52.38) (18.09, 75.25)
#> Partial Response (PR) 6 (40.0%) 5 (33.3%) 4 (26.7%)
#> 95% CI (Wald, with correction) (11.87, 68.13) (6.14, 60.52) (0.95, 52.38)
#> Stable Disease (SD) 5 (33.3%) 6 (40.0%) 4 (26.7%)
#> 95% CI (Wald, with correction) (6.14, 60.52) (11.87, 68.13) (0.95, 52.38)2. Best Overall Response (Ordering of treatment groups)
- By default, the first level or value of
arm_var(default to"ADSL.ARM"unless specified) is treated as the reference group without specification. - To apply user-defined reference group, please provide the value from
the treatment variable to the argument
ref_group, e.g.,ref_group = "PLACEBO". - Since
rtablesdisplays the reference group at the very left column, the order of displayed treatment groups may not be exactly the same as the order factorized, depending on which group is selected as the reference group. See below for examples:
Factorized trt order |
ref_group |
Displayed trt order |
Reference group used in analysis |
|---|---|---|---|
| ARM C, ARM B, ARM A | NULL | ARM C, ARM B, ARM A | ARM C |
| NULL | ARM B | ARM B, ARM A, ARM C | ARM B |
| ARM C, ARM B, ARM A | ARM B | ARM B, ARM C, ARM A | ARM B |
3. Best Overall Response (selecting sections to display)
- The section of
Odds Ratiocan be suppressed with the argumentodds_ratio = FALSE. - The section of
Difference in response ratecan be suppressed with the argumentperform_analysis = NULL.
proc_data <- log_filter(syn_data, PARAMCD == "BESRSPI", "adrs")
run(rspt01, proc_data, odds_ratio = FALSE, perform_analysis = NULL)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————
#> Responders 10 (66.7%) 9 (60.0%) 11 (73.3%)
#> 95% CI (Wald, with correction) (39.5, 93.9) (31.9, 88.1) (47.6, 99.0)
#> Complete Response (CR) 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> 95% CI (Wald, with correction) (0.95, 52.38) (0.95, 52.38) (18.09, 75.25)
#> Partial Response (PR) 6 (40.0%) 5 (33.3%) 4 (26.7%)
#> 95% CI (Wald, with correction) (11.87, 68.13) (6.14, 60.52) (0.95, 52.38)
#> Stable Disease (SD) 5 (33.3%) 6 (40.0%) 4 (26.7%)
#> 95% CI (Wald, with correction) (6.14, 60.52) (11.87, 68.13) (0.95, 52.38)4. Best Overall Response (with stratified analysis)
- A stratified analysis can be added by specifying the argument
perform_analysis = "strat"and providing the stratification variable to the argumentstrata. The argumentstratais expected ifperform_analysisis set to include stratified analysis. - The stratification variables are expected to be available in
adrs. - If both unstratified and stratified analysis are required, use
perform_analysis = c("unstrat", "strat")
proc_data <- log_filter(syn_data, PARAMCD == "BESRSPI", "adrs")
run(rspt01, proc_data, perform_analysis = "strat", strata = c("STRATA1", "STRATA2"))
#> Warning in prop_diff_cmh(rsp, grp, strata, conf_level): Less than 5
#> observations in some strata.
#> Warning in prop_diff_cmh(rsp, grp, strata, conf_level): Less than 5
#> observations in some strata.
#> Warning in prop_cmh(tbl): <5 data points in some strata. CMH test may be
#> incorrect.
#> Warning in prop_cmh(tbl): <5 data points in some strata. CMH test may be
#> incorrect.
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————————————————————
#> Responders 10 (66.7%) 9 (60.0%) 11 (73.3%)
#> 95% CI (Wald, with correction) (39.5, 93.9) (31.9, 88.1) (47.6, 99.0)
#> Stratified Analysis
#> Difference in Response rate (%) -11.0 22.5
#> 95% CI (CMH, without correction) (-42.7, 20.7) (-3.5, 48.5)
#> p-value (Cochran-Mantel-Haenszel Test) 0.5731 0.3088
#> Odds Ratio (95% CI) 0.75 (0.17 - 3.33) 1.37 (0.29 - 6.60)
#> Complete Response (CR) 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> 95% CI (Wald, with correction) (0.95, 52.38) (0.95, 52.38) (18.09, 75.25)
#> Partial Response (PR) 6 (40.0%) 5 (33.3%) 4 (26.7%)
#> 95% CI (Wald, with correction) (11.87, 68.13) (6.14, 60.52) (0.95, 52.38)
#> Stable Disease (SD) 5 (33.3%) 6 (40.0%) 4 (26.7%)
#> 95% CI (Wald, with correction) (6.14, 60.52) (11.87, 68.13) (0.95, 52.38)5. Best Overall Response (modifying analysis details like type of confidence interval, alpha level, test for p-value)
- The level of the confidence intervals is defined by the argument
conf_level. - The methods to construct confidence interval and p-value are
controlled by the argument
methods. It is a named list with five optional sub-arguments. For example,methods = list(prop_conf_method = "wald", diff_conf_method = "wald", strat_diff_conf_method = "ha", diff_pval_method = "fisher", strat_diff_pval_method = "schouten")
See table below for what each argument controls and the available method options:
| Arguments | Methods Controlled | Methods Options |
|---|---|---|
prop_conf_method |
proportion confidence interval |
"waldcc" (default), "wald",
etc. |
diff_conf_method |
unstratified difference confidence interval |
"waldcc" (default), "wald",
etc. |
diff_pval_method |
unstratified p-value for odds ratio |
"chisq" (default),
"fisher"
|
strat_diff_conf_method |
stratified difference confidence interval |
"cmh" (default), "ha"
|
strat_diff_pval_method |
stratified p-value for odds ratio |
"cmh" (default),
"schouten"
|
See in the table below the method options for estimates of proportions and the associated statistical methods:
| Method Options | Statistical Methods |
|---|---|
"clopper-pearson" |
Clopper-Pearson |
"wald" |
Wald, without correction |
"waldcc" |
Wald, with correction |
"wilson" |
Wilson, without correction |
"strat_wilson" |
Stratified Wilson, without correction |
"wilsonc" |
Wilson, with correction |
"strat_wilsonc" |
Stratified Wilson, with correction |
"agresti-coull" |
Agresti-Coull |
"jeffreys" |
Jeffreys |
See in the table below the method options for estimates of proportion difference and the associated statistical methods:
| Method Options | Statistical Methods |
|---|---|
"cmh" |
CMH, without correction |
"wald" |
Wald, with correction |
"waldcc" |
Wald, without correction |
"ha" |
Anderson-Hauck |
"newcombe" |
Newcombe, without correction |
"newcombecc" |
Newcombe, with correction |
"strat_wilsonc" |
Stratified Wilson, with correction |
"strat_newcombe" |
Stratified Newcombe, without correction |
"strat_newcombecc" |
Stratified Newcombe, with correction |
See in the table below the method options for testing proportion difference and the associated statistical methods:
| Method Options | Statistical Methods |
|---|---|
"chisq" |
Chi-Squared test |
"fisher" |
the Fisher’s exact test |
"cmh" |
stratified Cochran-Mantel-Haenszel test |
"shouten" |
Chi-Squared test with Schouten correction |
An example:
proc_data <- log_filter(syn_data, PARAMCD == "BESRSPI", "adrs")
run(rspt01, proc_data,
conf_level = 0.90,
methods = list(
prop_conf_method = "wald",
diff_conf_method = "wald",
diff_pval_method = "fisher"
)
)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————————————————
#> Responders 10 (66.7%) 9 (60.0%) 11 (73.3%)
#> 90% CI (Wald, without correction) (46.6, 86.7) (39.2, 80.8) (54.6, 92.1)
#> Unstratified Analysis
#> Difference in Response rate (%) -6.7 6.7
#> 90% CI (Wald, without correction) (-35.5, 22.2) (-20.8, 34.1)
#> p-value (Fisher's Exact Test) 1.0000 1.0000
#> Odds Ratio (95% CI) 0.75 (0.17 - 3.33) 1.37 (0.29 - 6.60)
#> Complete Response (CR) 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> 90% CI (Wald, without correction) (7.89, 45.45) (7.89, 45.45) (25.48, 67.85)
#> Partial Response (PR) 6 (40.0%) 5 (33.3%) 4 (26.7%)
#> 90% CI (Wald, without correction) (19.19, 60.81) (13.31, 53.35) (7.89, 45.45)
#> Stable Disease (SD) 5 (33.3%) 6 (40.0%) 4 (26.7%)
#> 90% CI (Wald, without correction) (13.31, 53.35) (19.19, 60.81) (7.89, 45.45)6. Best Overall Response (modifying the definition of overall response)
The following example shows how to customize the definition of responder, e.g, consider only complete response as response.
proc_data <- log_filter(syn_data, PARAMCD == "BESRSPI", "adrs")
preprocess(rspt01) <- function(adam_db, ...) {
adam_db$adrs <- adam_db$adrs %>%
mutate(RSP_LAB = tern::d_onco_rsp_label(.data$AVALC)) %>%
mutate(IS_RSP = .data$AVALC %in% c("CR"))
adam_db
}
run(rspt01, proc_data)
#> Warning in stats::prop.test(tbl, correct = FALSE): Chi-squared approximation
#> may be incorrect
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ——————————————————————————————————————————————————————————————————————————————————————————————
#> Responders 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> 95% CI (Wald, with correction) (1.0, 52.4) (1.0, 52.4) (18.1, 75.2)
#> Unstratified Analysis
#> Difference in Response rate (%) 0.0 20.0
#> 95% CI (Wald, with correction) (-38.3, 38.3) (-20.4, 60.4)
#> p-value (Chi-Squared Test) 1.0000 0.2557
#> Odds Ratio (95% CI) 1.00 (0.20 - 5.04) 2.41 (0.52 - 11.10)
#> Complete Response (CR) 4 (26.7%) 4 (26.7%) 7 (46.7%)
#> 95% CI (Wald, with correction) (0.95, 52.38) (0.95, 52.38) (18.09, 75.25)
#> Partial Response (PR) 6 (40.0%) 5 (33.3%) 4 (26.7%)
#> 95% CI (Wald, with correction) (11.87, 68.13) (6.14, 60.52) (0.95, 52.38)
#> Stable Disease (SD) 5 (33.3%) 6 (40.0%) 4 (26.7%)
#> 95% CI (Wald, with correction) (6.14, 60.52) (11.87, 68.13) (0.95, 52.38)
Time-to-event Summary (TTET01)
1. Time-to-event Summary
- The
ttet01template produces the standard time-to-event summary. - Users are expected to subset the parameter of interest
(e.g.
PARAMCD == "PFS") in pre-processing. - Please see the section of Best Overall Response (Ordering of treatment groups) to find out more about the ordering of treatment groups and reference group.
- Unstratified analysis is provided by default.
- Survival estimations and difference in survival are both provided by default.
proc_data <- log_filter(syn_data, PARAMCD == "PFS", "adtte")
run(ttet01, proc_data)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————
#> Patients with event (%) 7 (46.7%) 12 (80%) 8 (53.3%)
#> Earliest contributing event
#> Death 5 11 7
#> Disease Progression 2 1 1
#> Patients without event (%) 8 (53.3%) 3 (20%) 7 (46.7%)
#> Time to Event (MONTHS)
#> Median 8.6 6.2 8.4
#> 95% CI (7.3, NE) (4.8, 7.6) (7.0, NE)
#> 25% and 75%-ile 3.8, NE 4.7, 8.4 5.8, NE
#> Range 1.2 to 9.5 {1} 0.9 to 9.1 0.9 to 9.5 {1}
#> Unstratified Analysis
#> p-value (log-rank) 0.0973 0.9111
#> Hazard Ratio 2.18 1.06
#> 95% CI (0.85, 5.60) (0.38, 2.94)
#> 6 MONTHS
#> Patients remaining at risk 11 8 11
#> Event Free Rate (%) 73.33 53.33 73.33
#> 95% CI (50.95, 95.71) (28.09, 78.58) (50.95, 95.71)
#> Difference in Event Free Rate -20.00 0.00
#> 95% CI (-53.74, 13.74) (-31.65, 31.65)
#> p-value (Z-test) 0.2453 1.0000
#> ————————————————————————————————————————————————————————————————————————————————————
#>
#> {1} - Censored observation: range maximum
#> ————————————————————————————————————————————————————————————————————————————————————2. Time-to-event Summary (selecting sections to display)
To suspend the section of earliest contributing events, use
summarize_event = FALSE.
proc_data <- log_filter(syn_data, PARAMCD == "PFS", "adtte")
run(ttet01, proc_data, summarize_event = FALSE)
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————
#> Patients with event (%) 7 (46.7%) 12 (80%) 8 (53.3%)
#> Patients without event (%) 8 (53.3%) 3 (20%) 7 (46.7%)
#> Time to Event (MONTHS)
#> Median 8.6 6.2 8.4
#> 95% CI (7.3, NE) (4.8, 7.6) (7.0, NE)
#> 25% and 75%-ile 3.8, NE 4.7, 8.4 5.8, NE
#> Range 1.2 to 9.5 {1} 0.9 to 9.1 0.9 to 9.5 {1}
#> Unstratified Analysis
#> p-value (log-rank) 0.0973 0.9111
#> Hazard Ratio 2.18 1.06
#> 95% CI (0.85, 5.60) (0.38, 2.94)
#> 6 MONTHS
#> Patients remaining at risk 11 8 11
#> Event Free Rate (%) 73.33 53.33 73.33
#> 95% CI (50.95, 95.71) (28.09, 78.58) (50.95, 95.71)
#> Difference in Event Free Rate -20.00 0.00
#> 95% CI (-53.74, 13.74) (-31.65, 31.65)
#> p-value (Z-test) 0.2453 1.0000
#> ————————————————————————————————————————————————————————————————————————————————————
#>
#> {1} - Censored observation: range maximum
#> ————————————————————————————————————————————————————————————————————————————————————To select either survival estimations or difference in survival or
both, please specify in the argument method. -
surv calls out the analysis of patients remaining at risk,
event free rate and corresponding 95% confidence interval of the rates.
- surv_diff calls out the analysis of difference in event
free rate, the 95% confidence interval of the difference and its
corresponding p-value. - both calls out both.
proc_data <- log_filter(syn_data, PARAMCD == "PFS", "adtte")
run(ttet01, proc_data, method = "surv")
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————
#> Patients with event (%) 7 (46.7%) 12 (80%) 8 (53.3%)
#> Earliest contributing event
#> Death 5 11 7
#> Disease Progression 2 1 1
#> Patients without event (%) 8 (53.3%) 3 (20%) 7 (46.7%)
#> Time to Event (MONTHS)
#> Median 8.6 6.2 8.4
#> 95% CI (7.3, NE) (4.8, 7.6) (7.0, NE)
#> 25% and 75%-ile 3.8, NE 4.7, 8.4 5.8, NE
#> Range 1.2 to 9.5 {1} 0.9 to 9.1 0.9 to 9.5 {1}
#> Unstratified Analysis
#> p-value (log-rank) 0.0973 0.9111
#> Hazard Ratio 2.18 1.06
#> 95% CI (0.85, 5.60) (0.38, 2.94)
#> 6 MONTHS
#> Patients remaining at risk 11 8 11
#> Event Free Rate (%) 73.33 53.33 73.33
#> 95% CI (50.95, 95.71) (28.09, 78.58) (50.95, 95.71)
#> ————————————————————————————————————————————————————————————————————————————————
#>
#> {1} - Censored observation: range maximum
#> ————————————————————————————————————————————————————————————————————————————————3. Time-to-event Summary (modifying analysis details like confidence interval type, ties, and alpha level)
- The level of the confidence intervals is defined by the argument
conf_level. - The type of confidence interval is defined in the argument
conf_type. Options are"plain"(default),"log"and"log-log". - Handling of ties is specified in the argument
ties. Options are"efron"(default),"breslow"or"exact".
proc_data <- log_filter(syn_data, PARAMCD == "PFS", "adtte")
run(ttet01, proc_data, conf_level = 0.90, conf_type = "log-log", ties = "efron")
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————————————————
#> Patients with event (%) 7 (46.7%) 12 (80%) 8 (53.3%)
#> Earliest contributing event
#> Death 5 11 7
#> Disease Progression 2 1 1
#> Patients without event (%) 8 (53.3%) 3 (20%) 7 (46.7%)
#> Time to Event (MONTHS)
#> Median 8.6 6.2 8.4
#> 90% CI (3.8, NE) (4.7, 7.6) (5.8, NE)
#> 25% and 75%-ile 3.8, NE 4.7, 8.4 5.8, NE
#> Range 1.2 to 9.5 {1} 0.9 to 9.1 0.9 to 9.5 {1}
#> Unstratified Analysis
#> p-value (log-rank) 0.0973 0.9111
#> Hazard Ratio 2.18 1.06
#> 90% CI (0.99, 4.81) (0.45, 2.50)
#> 6 MONTHS
#> Patients remaining at risk 11 8 11
#> Event Free Rate (%) 73.33 53.33 73.33
#> 90% CI (49.25, 87.30) (30.65, 71.60) (49.25, 87.30)
#> Difference in Event Free Rate -20.00 0.00
#> 90% CI (-48.31, 8.31) (-26.56, 26.56)
#> p-value (Z-test) 0.2453 1.0000
#> ———————————————————————————————————————————————————————————————————————————————————
#>
#> {1} - Censored observation: range maximum
#> ———————————————————————————————————————————————————————————————————————————————————4. Time-to-event Summary (with stratified analysis)
- A stratified analysis can be added by specifying the argument
perform_analysis = "strat"and providing the stratification variable to the argumentstrata. The argumentstratais expected ifperform_analysisis set to include stratified analysis. - The stratification variables are expected to be available in
adrs. - If unstratified and stratified analysis are both required, users can
use
perform_analysis = c("unstrat", "strat").
proc_data <- log_filter(syn_data, PARAMCD == "PFS", "adtte")
run(ttet01, proc_data, perform_analysis = "strat", strata = "STRATA1")
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————
#> Patients with event (%) 7 (46.7%) 12 (80%) 8 (53.3%)
#> Earliest contributing event
#> Death 5 11 7
#> Disease Progression 2 1 1
#> Patients without event (%) 8 (53.3%) 3 (20%) 7 (46.7%)
#> Time to Event (MONTHS)
#> Median 8.6 6.2 8.4
#> 95% CI (7.3, NE) (4.8, 7.6) (7.0, NE)
#> 25% and 75%-ile 3.8, NE 4.7, 8.4 5.8, NE
#> Range 1.2 to 9.5 {1} 0.9 to 9.1 0.9 to 9.5 {1}
#> Stratified Analysis
#> p-value (log-rank) 0.0649 0.8901
#> Hazard Ratio 2.52 1.08
#> 95% CI (0.92, 6.93) (0.36, 3.22)
#> 6 MONTHS
#> Patients remaining at risk 11 8 11
#> Event Free Rate (%) 73.33 53.33 73.33
#> 95% CI (50.95, 95.71) (28.09, 78.58) (50.95, 95.71)
#> Difference in Event Free Rate -20.00 0.00
#> 95% CI (-53.74, 13.74) (-31.65, 31.65)
#> p-value (Z-test) 0.2453 1.0000
#> ————————————————————————————————————————————————————————————————————————————————————
#>
#> {1} - Censored observation: range maximum
#> ————————————————————————————————————————————————————————————————————————————————————5. Time-to-event Summary (modifying time point for the “survival at xx months” analysis)
The time point for the “survival at xx months” analysis can be
modified by specifying the argument time_point. By default,
the function takes AVAL from adtte in days and
converts it to months. The survival estimates are then summarized in
month, and the numeric values should be provided in months to
time_point.
proc_data <- log_filter(syn_data, PARAMCD == "PFS", "adtte")
run(ttet01, proc_data, perform_analysis = "unstrat", time_point = c(3, 6))
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————
#> Patients with event (%) 7 (46.7%) 12 (80%) 8 (53.3%)
#> Earliest contributing event
#> Death 5 11 7
#> Disease Progression 2 1 1
#> Patients without event (%) 8 (53.3%) 3 (20%) 7 (46.7%)
#> Time to Event (MONTHS)
#> Median 8.6 6.2 8.4
#> 95% CI (7.3, NE) (4.8, 7.6) (7.0, NE)
#> 25% and 75%-ile 3.8, NE 4.7, 8.4 5.8, NE
#> Range 1.2 to 9.5 {1} 0.9 to 9.1 0.9 to 9.5 {1}
#> Unstratified Analysis
#> p-value (log-rank) 0.0973 0.9111
#> Hazard Ratio 2.18 1.06
#> 95% CI (0.85, 5.60) (0.38, 2.94)
#> 3 MONTHS
#> Patients remaining at risk 12 12 13
#> Event Free Rate (%) 80.00 80.00 86.67
#> 95% CI (59.76, 100.00) (59.76, 100.00) (69.46, 100.00)
#> Difference in Event Free Rate 0.00 6.67
#> 95% CI (-28.63, 28.63) (-19.90, 33.23)
#> p-value (Z-test) 1.0000 0.6228
#> 6 MONTHS
#> Patients remaining at risk 11 8 11
#> Event Free Rate (%) 73.33 53.33 73.33
#> 95% CI (50.95, 95.71) (28.09, 78.58) (50.95, 95.71)
#> Difference in Event Free Rate -20.00 0.00
#> 95% CI (-53.74, 13.74) (-31.65, 31.65)
#> p-value (Z-test) 0.2453 1.0000
#> —————————————————————————————————————————————————————————————————————————————————————
#>
#> {1} - Censored observation: range maximum
#> —————————————————————————————————————————————————————————————————————————————————————The following example shows how to specify the time point in user-defined unit.
proc_data <- log_filter(syn_data, PARAMCD == "PFS", "adtte")
preprocess(ttet01) <- function(adam_db, dataset = "adtte",
...) {
adam_db[[dataset]] <- adam_db[[dataset]] %>%
mutate(
AVALU = "DAYS",
IS_EVENT = .data$CNSR == 0,
IS_NOT_EVENT = .data$CNSR == 1,
EVNT1 = factor(
case_when(
IS_EVENT == TRUE ~ render_safe("{Patient_label} with event (%)"),
IS_EVENT == FALSE ~ render_safe("{Patient_label} without event (%)")
),
levels = render_safe(c("{Patient_label} with event (%)", "{Patient_label} without event (%)"))
),
EVNTDESC = factor(.data$EVNTDESC)
)
adam_db
}
run(ttet01, proc_data, perform_analysis = "unstrat", time_point = c(91, 183))
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————————————————————
#> Patients with event (%) 7 (46.7%) 12 (80%) 8 (53.3%)
#> Earliest contributing event
#> Death 5 11 7
#> Disease Progression 2 1 1
#> Patients without event (%) 8 (53.3%) 3 (20%) 7 (46.7%)
#> Time to Event (DAYS)
#> Median 261.9 187.7 256.3
#> 95% CI (221.9, NE) (144.7, 232.2) (212.0, NE)
#> 25% and 75%-ile 114.9, NE 141.9, 254.4 175.0, NE
#> Range 37.2 to 288.3 {1} 28.0 to 276.6 26.4 to 288.1 {1}
#> Unstratified Analysis
#> p-value (log-rank) 0.0973 0.9111
#> Hazard Ratio 2.18 1.06
#> 95% CI (0.85, 5.60) (0.38, 2.94)
#> 91 DAYS
#> Patients remaining at risk 12 12 13
#> Event Free Rate (%) 80.00 80.00 86.67
#> 95% CI (59.76, 100.00) (59.76, 100.00) (69.46, 100.00)
#> Difference in Event Free Rate 0.00 6.67
#> 95% CI (-28.63, 28.63) (-19.90, 33.23)
#> p-value (Z-test) 1.0000 0.6228
#> 183 DAYS
#> Patients remaining at risk 11 8 11
#> Event Free Rate (%) 73.33 53.33 73.33
#> 95% CI (50.95, 95.71) (28.09, 78.58) (50.95, 95.71)
#> Difference in Event Free Rate -20.00 0.00
#> 95% CI (-53.74, 13.74) (-31.65, 31.65)
#> p-value (Z-test) 0.2453 1.0000
#> —————————————————————————————————————————————————————————————————————————————————————————
#>
#> {1} - Censored observation: range maximum
#> —————————————————————————————————————————————————————————————————————————————————————————6. Time-to-event Summary (modifying the p-value method for testing hazard ratio)
The default p-value method for testing hazard ratio is “log-rank”.
Alternative methods can be requested by specifying the argument
pval_method and options include, log-rank
(default), wald or likelihood. The syntax
currently does not allow requesting more than one p-value.
Note that ttet01 has been modified in the previous
example (i.e., preprocess(ttet01) has been overridden); to
access the default template, try chevron::ttet01.
proc_data <- log_filter(syn_data, PARAMCD == "PFS", "adtte")
run(ttet01, proc_data, pval_method = "wald")
#> A: Drug X B: Placebo C: Combination
#> (N=15) (N=15) (N=15)
#> ————————————————————————————————————————————————————————————————————————————————————————
#> Patients with event (%) 7 (46.7%) 12 (80%) 8 (53.3%)
#> Earliest contributing event
#> Death 5 11 7
#> Disease Progression 2 1 1
#> Patients without event (%) 8 (53.3%) 3 (20%) 7 (46.7%)
#> Time to Event (DAYS)
#> Median 261.9 187.7 256.3
#> 95% CI (221.9, NE) (144.7, 232.2) (212.0, NE)
#> 25% and 75%-ile 114.9, NE 141.9, 254.4 175.0, NE
#> Range 37.2 to 288.3 {1} 28.0 to 276.6 26.4 to 288.1 {1}
#> Unstratified Analysis
#> p-value (wald) 0.1053 0.9111
#> Hazard Ratio 2.18 1.06
#> 95% CI (0.85, 5.60) (0.38, 2.94)
#> 6 DAYS
#> Patients remaining at risk 15 15 15
#> Event Free Rate (%) 100.00 100.00 100.00
#> 95% CI (100.00, 100.00) (100.00, 100.00) (100.00, 100.00)
#> 12 DAYS
#> Patients remaining at risk 15 15 15
#> Event Free Rate (%) 100.00 100.00 100.00
#> 95% CI (100.00, 100.00) (100.00, 100.00) (100.00, 100.00)
#> ————————————————————————————————————————————————————————————————————————————————————————
#>
#> {1} - Censored observation: range maximum
#> ————————————————————————————————————————————————————————————————————————————————————————
Vital Signs (VST01)
1. Vital Sign Results and Change from Baseline by Visit
t_vs_chg <- run(vst01, syn_data)
head(t_vs_chg, 20)
#> A: Drug X B: Placebo C: Combination
#> Change from Change from Change from
#> Value at Visit Baseline Value at Visit Baseline Value at Visit Baseline
#> (N=15) (N=15) (N=15) (N=15) (N=15) (N=15)
#> ——————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————————
#> Diastolic Blood Pressure
#> SCREENING
#> n 15 0 15 0 15 0
#> Mean (SD) 94.385 (17.067) NE (NE) 106.381 (20.586) NE (NE) 106.468 (12.628) NE (NE)
#> Median 94.933 NE 111.133 NE 108.359 NE
#> Min - Max 55.71 - 122.00 NE - NE 60.21 - 131.91 NE - NE 83.29 - 127.17 NE - NE
#> BASELINE
#> n 15 15 15
#> Mean (SD) 96.133 (22.458) 108.111 (15.074) 103.149 (19.752)
#> Median 93.328 108.951 102.849
#> Min - Max 60.58 - 136.59 83.44 - 131.62 66.05 - 136.55
#> WEEK 1 DAY 8
#> n 15 15 15 15 15 15
#> Mean (SD) 98.977 (21.359) 2.844 (28.106) 104.110 (16.172) -4.001 (21.867) 100.826 (19.027) -2.323 (25.018)
#> Median 92.447 -4.066 107.703 3.227 103.058 -2.476
#> Min - Max 67.55 - 130.37 -32.82 - 47.68 70.91 - 132.89 -52.94 - 28.63 70.04 - 128.68 -55.15 - 41.81
#> WEEK 2 DAY 15
#> n 15 15 15 15 15 15
#> Mean (SD) 99.758 (14.477) 3.626 (21.189) 97.473 (17.296) -10.638 (20.831) 94.272 (16.961) -8.877 (27.229)
#> Median 101.498 1.731 99.501 -9.727 96.789 -10.155
Vital Signs Abnormalities (Regardless of Abnormality at
Baseline) (VST02_1)
1. Vital Sign Abnormalities (Regardless of Abnormality at Baseline)
run(vst02_1, syn_data)
#> Assessment A: Drug X B: Placebo C: Combination
#> Abnormality (N=15) (N=15) (N=15)
#> —————————————————————————————————————————————————————————————————————————
#> Diastolic Blood Pressure
#> Low 8/15 (53.3%) 9/15 (60%) 8/15 (53.3%)
#> High 10/15 (66.7%) 5/15 (33.3%) 8/15 (53.3%)
#> Pulse Rate
#> Low 9/15 (60%) 3/15 (20%) 5/15 (33.3%)
#> High 2/15 (13.3%) 6/15 (40%) 5/15 (33.3%)
#> Respiratory Rate
#> Low 13/15 (86.7%) 10/15 (66.7%) 13/15 (86.7%)
#> High 7/15 (46.7%) 10/15 (66.7%) 11/15 (73.3%)
#> Systolic Blood Pressure
#> Low 7/15 (46.7%) 9/15 (60%) 11/15 (73.3%)
#> High 10/15 (66.7%) 9/15 (60%) 9/15 (60%)
#> Temperature
#> Low 12/15 (80%) 13/15 (86.7%) 11/15 (73.3%)
#> High 14/15 (93.3%) 12/15 (80%) 14/15 (93.3%)
#> Weight
#> Low 3/15 (20%) 3/15 (20%) 4/15 (26.7%)
#> High 4/15 (26.7%) 4/15 (26.7%) 5/15 (33.3%)
Vital Signs Abnormalities (Among Subject Without Abnormality
at Baseline) (VST02_2)
1. Vital Sign Abnormalities (Among Subject Without Abnormality at Baseline)
run(vst02_2, syn_data)
#> Assessment A: Drug X B: Placebo C: Combination
#> Abnormality (N=15) (N=15) (N=15)
#> ———————————————————————————————————————————————————————————————————————
#> Diastolic Blood Pressure
#> Low 6/11 (54.5%) 9/15 (60%) 6/12 (50%)
#> High 8/12 (66.7%) 4/11 (36.4%) 7/13 (53.8%)
#> Pulse Rate
#> Low 9/15 (60%) 3/15 (20%) 5/13 (38.5%)
#> High 2/14 (14.3%) 4/12 (33.3%) 5/15 (33.3%)
#> Respiratory Rate
#> Low 7/9 (77.8%) 7/11 (63.6%) 11/12 (91.7%)
#> High 6/14 (42.9%) 7/11 (63.6%) 9/13 (69.2%)
#> Systolic Blood Pressure
#> Low 5/13 (38.5%) 8/12 (66.7%) 10/14 (71.4%)
#> High 8/13 (61.5%) 8/13 (61.5%) 8/13 (61.5%)
#> Temperature
#> Low 8/10 (80%) 7/9 (77.8%) 8/10 (80%)
#> High 8/8 (100%) 7/8 (87.5%) 12/13 (92.3%)
#> Weight
#> Low 3/15 (20%) 3/15 (20%) 3/14 (21.4%)
#> High 4/14 (28.6%) 4/15 (26.7%) 5/14 (35.7%)LISTINGS
Glossary of Adverse Event Preferred Terms and
Investigator-Specified Terms (AEL01_NOLLT)
1. Glossary of Adverse Event Preferred Terms and Investigator-Specified Terms
- The
ael01_nollttemplate produces the standard glossary of adverse event preferred terms and investigator-specified terms. - The example below uses
headfunction to print only the first 10 lines of the output.
l_ae_nollt <- run(ael01_nollt, syn_data)
head(l_ae_nollt, 10)
#> MedDRA System Organ Class MedDRA Preferred Term Reported Term for the Adverse Event
#> ———————————————————————————————————————————————————————————————————————————————————————
#> cl A.1 dcd A.1.1.1.1 trm A.1.1.1.1
#> dcd A.1.1.1.2 trm A.1.1.1.2
#> cl B.1 dcd B.1.1.1.1 trm B.1.1.1.1
#> cl B.2 dcd B.2.1.2.1 trm B.2.1.2.1
#> dcd B.2.2.3.1 trm B.2.2.3.1
#> cl C.1 dcd C.1.1.1.3 trm C.1.1.1.3
#> cl C.2 dcd C.2.1.2.1 trm C.2.1.2.1
#> cl D.1 dcd D.1.1.1.1 trm D.1.1.1.1
#> dcd D.1.1.4.2 trm D.1.1.4.2
#> cl D.2 dcd D.2.1.5.3 trm D.2.1.5.3Graphics
Forest Plot for Odds Ratio
(FSTG01)
1. Forest Plot for Odds Ratio (with subgroup analysis)
- The
fstg01template produces the standard forest plot for odds ratio. - Users are expected to subset the parameter of interest
(e.g.
PARAMCD == "BESRSPI") in pre-processing. - Users are expected to subset the arm variable to keep only the two
arms to compare
(e.g.
ARM %in% c("A: Drug X", "B: Placebo")). - By default, the plots displays a subgroup analysis for
"SEX","AGEGR1"and"RACE". - Unstratified analysis is provided by default.
- The plots displays by default the Total number of subjects, the odd ratio and the 95% confidence interval, and, for each arm, the number of subject, the number of responders and the proportion of responders.
proc_data <- log_filter(
syn_data,
PARAMCD == "BESRSPI" & ARM %in% c("A: Drug X", "B: Placebo"), "adrs"
)
run(fstg01, proc_data)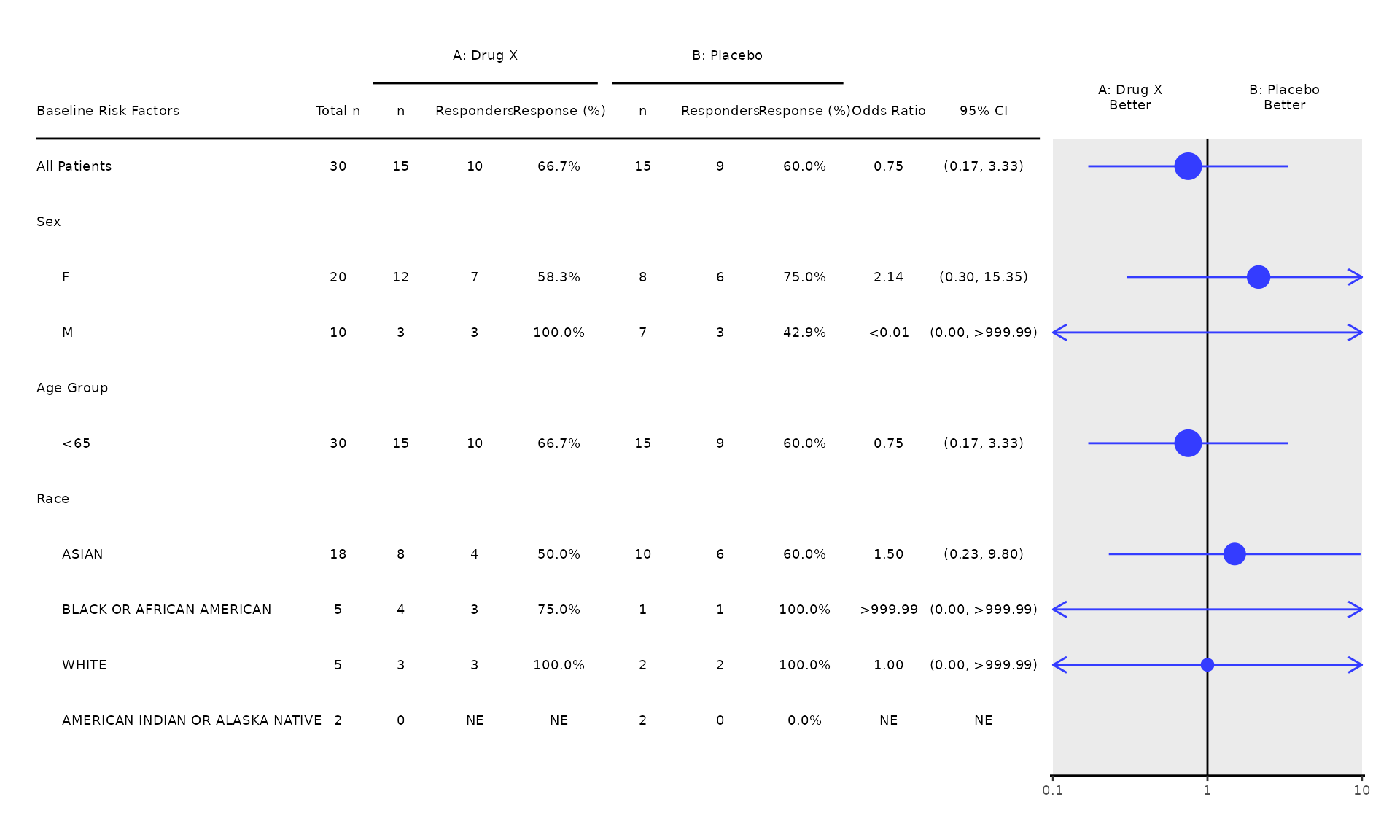
2. Forest Plot for Odds Ratio (with a user-defined confidence level)
The confidence level of the confidence interval can be adjusted by
the conf_level argument.
run(fstg01, proc_data, conf_level = 0.90)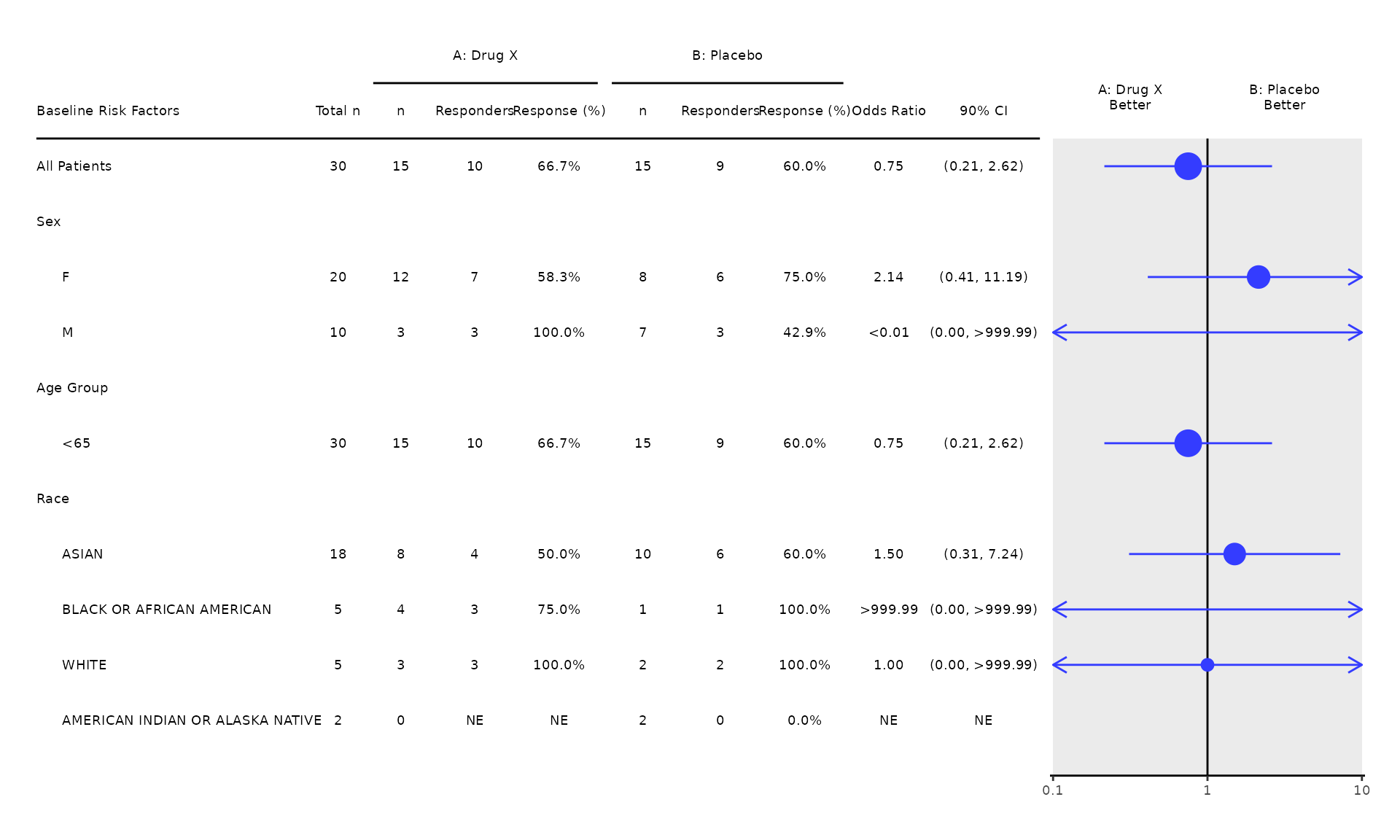
3. Forest Plot for Odds Ratio (with p-values and/or different statistics)
The interaction p-values and a different set of statistics can be
displayed using the stat_var argument. Note that the users
are expected to select a method for p-value computation. see
[tern::prop_diff_test].
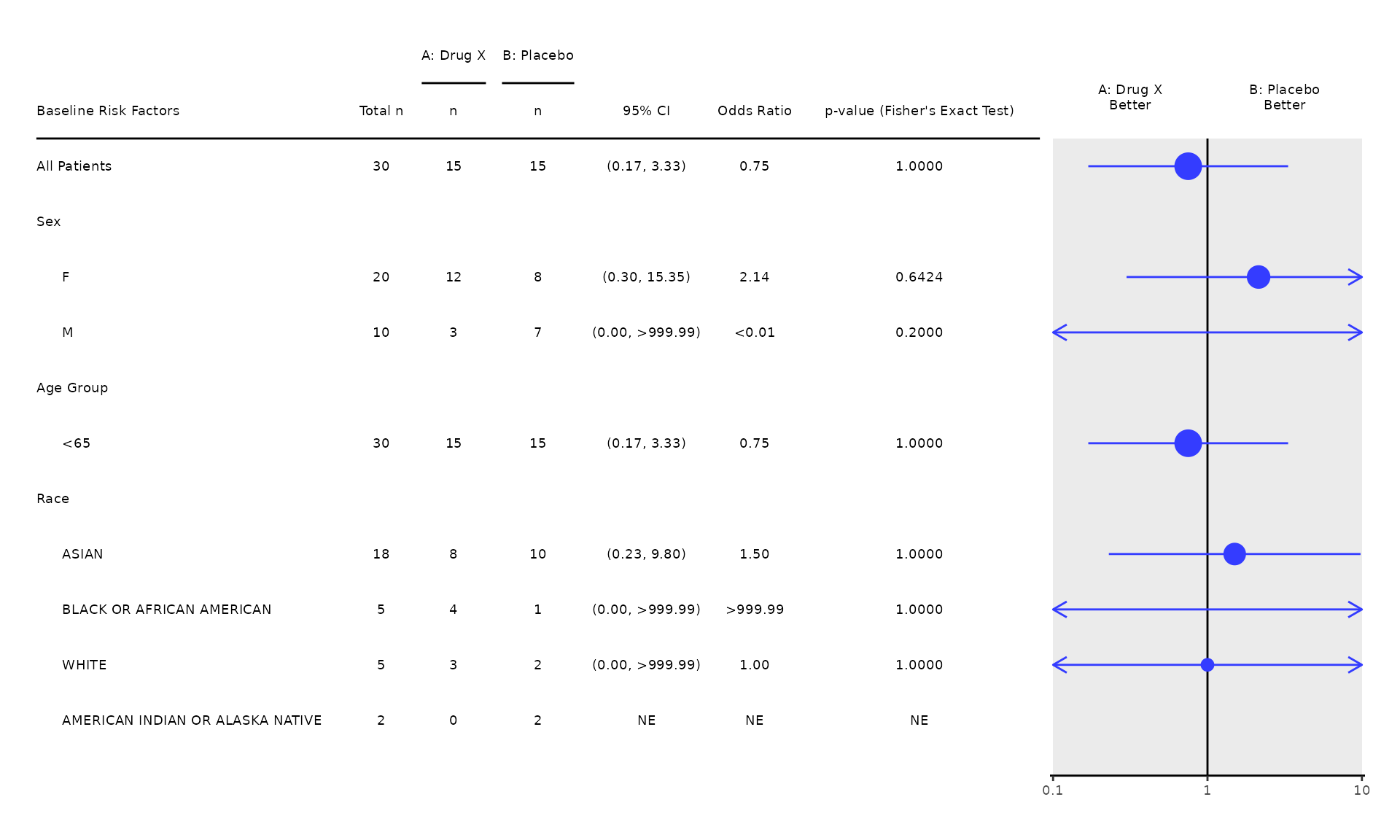
4. Forest Plot for Odds Ratio (with user-defined subgroup analysis)
The subgroups arguments controls which variables are
used for subgroup analysis. If NULLthe subgroup analysis is
removed.
run(fstg01, proc_data, subgroups = NULL)
5. Forest Plot for Odds Ratio (with stratified analysis)
The strata_var argument is used to pass the columns used
for stratified analysis.
run(fstg01, proc_data, strata_var = "STRATA1")
#> Warning in coxexact.fit(X, Y, istrat, offset, init, control, weights = weights,
#> : Ran out of iterations and did not converge
#> Warning in s_odds_ratio(df = l_df[[2]], .var = "rsp", .ref_group = l_df[[1]], :
#> Unable to compute the odds ratio estimate. Please try re-running the function
#> with parameter `method` set to "approximate".
#> Warning in coxexact.fit(X, Y, istrat, offset, init, control, weights = weights,
#> : Ran out of iterations and did not converge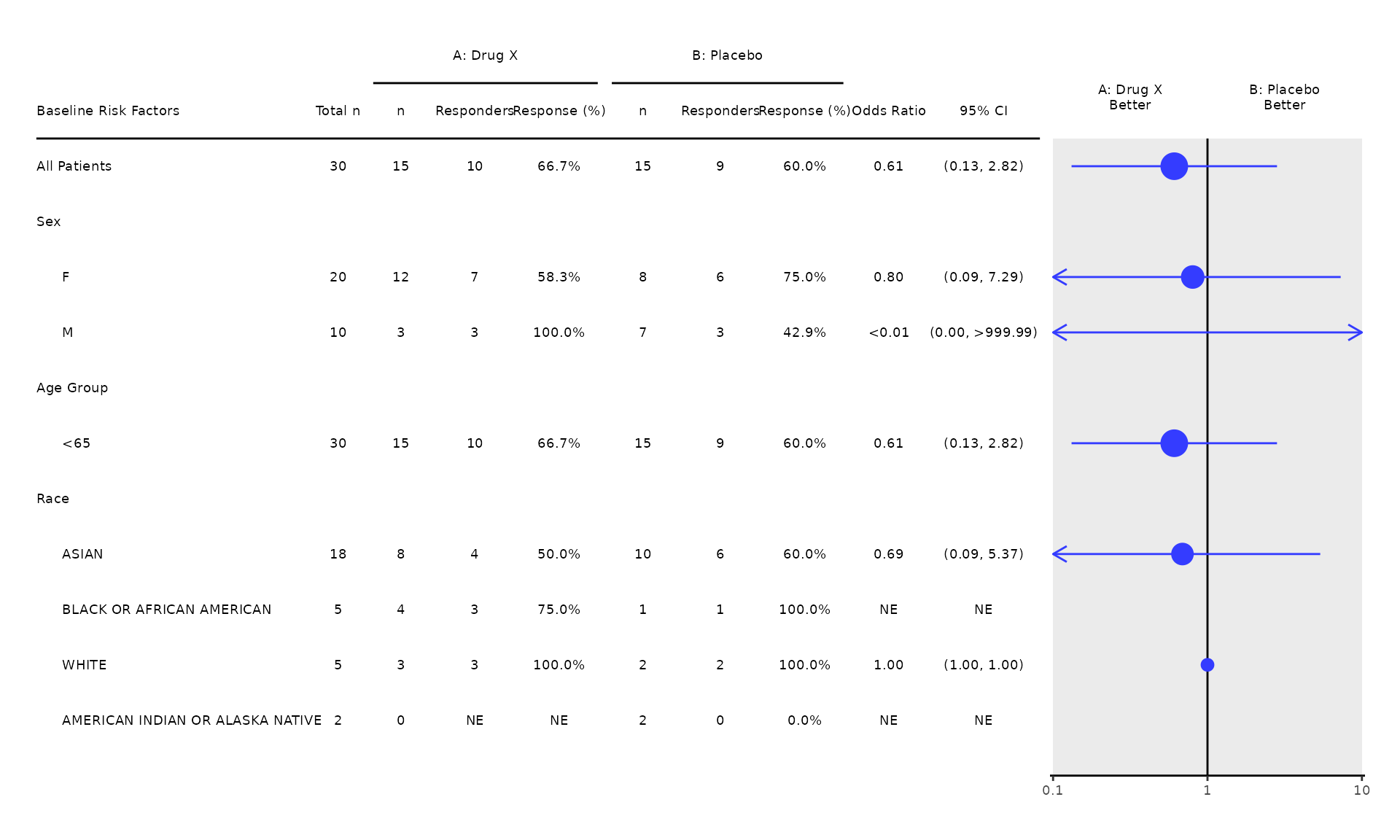
6. Forest Plot for Odds Ratio (without proportional sizing of the odds ratio symbol)
The col_symbol_size argument controls the size of the
odds ratio symbols which are by default proportional in size to the
sample size of the subgroup. If NULL the same symbol size
is used for all subgroups.
run(fstg01, proc_data, col_symbol_size = NULL)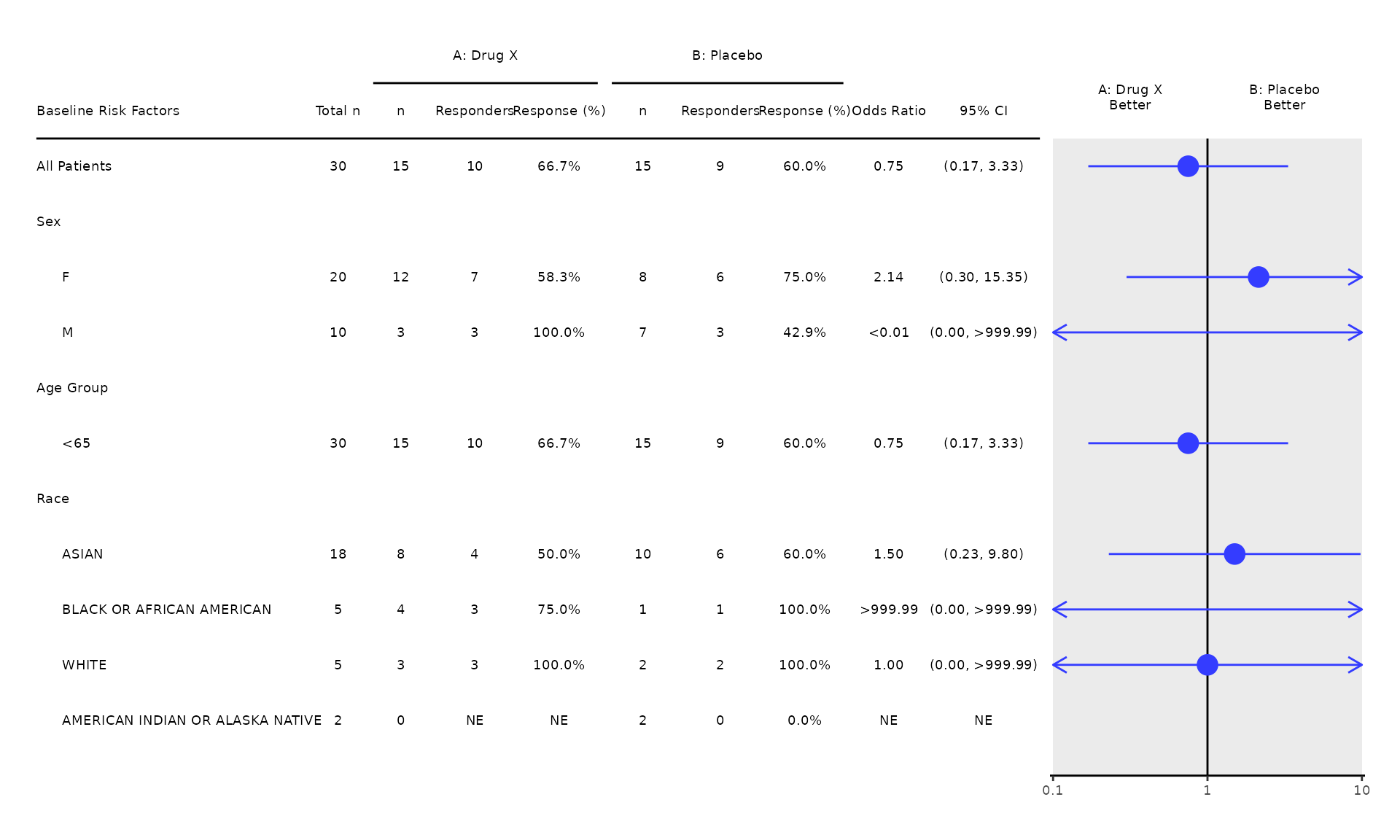
Forest Plot for Hazard Ratio
(FSTG02)
1. Forest Plot for Hazard Ratio (with subgroup analysis)
- The
fstg02template produces the standard forest plot for hazard ratio. - Users are expected to subset the parameter of interest
(e.g.
PARAMCD == "OS") in pre-processing. - Users are expected to subset the arm variable to keep only the two
arms to compare
(e.g.
ARM %in% c("A: Drug X", "B: Placebo")). - By default, the plots displays a subgroup analysis for
"SEX","AGEGR1"and"RACE". - Unstratified analysis is provided by default.
- The plots displays by default the Total number of events, the hazard ratio and the 95% confidence interval, and, for each arm, the number of events and the median time to event in month.
proc_data <- log_filter(
syn_data,
PARAMCD == "OS" & ARM %in% c("A: Drug X", "B: Placebo"), "adtte"
)
run(fstg02, proc_data)
2. Forest Plot for Hazard Ratio (with p-values and/or different statistics)
The interaction p-values and a different set of statistics can be
displayed using the control argument. More details about
the control options are available in
[tern::extract_survival_subgroups]
run(
fstg02,
proc_data,
stat_var = c("n_tot", "n", "ci", "hr", "pval"),
control = list(conf_level = 0.9, pval_method = "likelihood")
)
3. Forest Plot for Hazard Ratio (with user-defined subgroup analysis)
The subgroups arguments controls which variables are
used for subgroup analysis. If NULLthe subgroup analysis is
removed.
run(fstg02, proc_data, subgroups = NULL)
4. Forest Plot for Hazard Ratio (with stratified analysis)
The strata_var argument is used to pass the columns used
for stratified analysis.
run(fstg02, proc_data, strata_var = "STRATA1")
#> Warning in coxph.fit(X, Y, istrat, offset, init, control, weights = weights, :
#> Loglik converged before variable 1 ; coefficient may be infinite.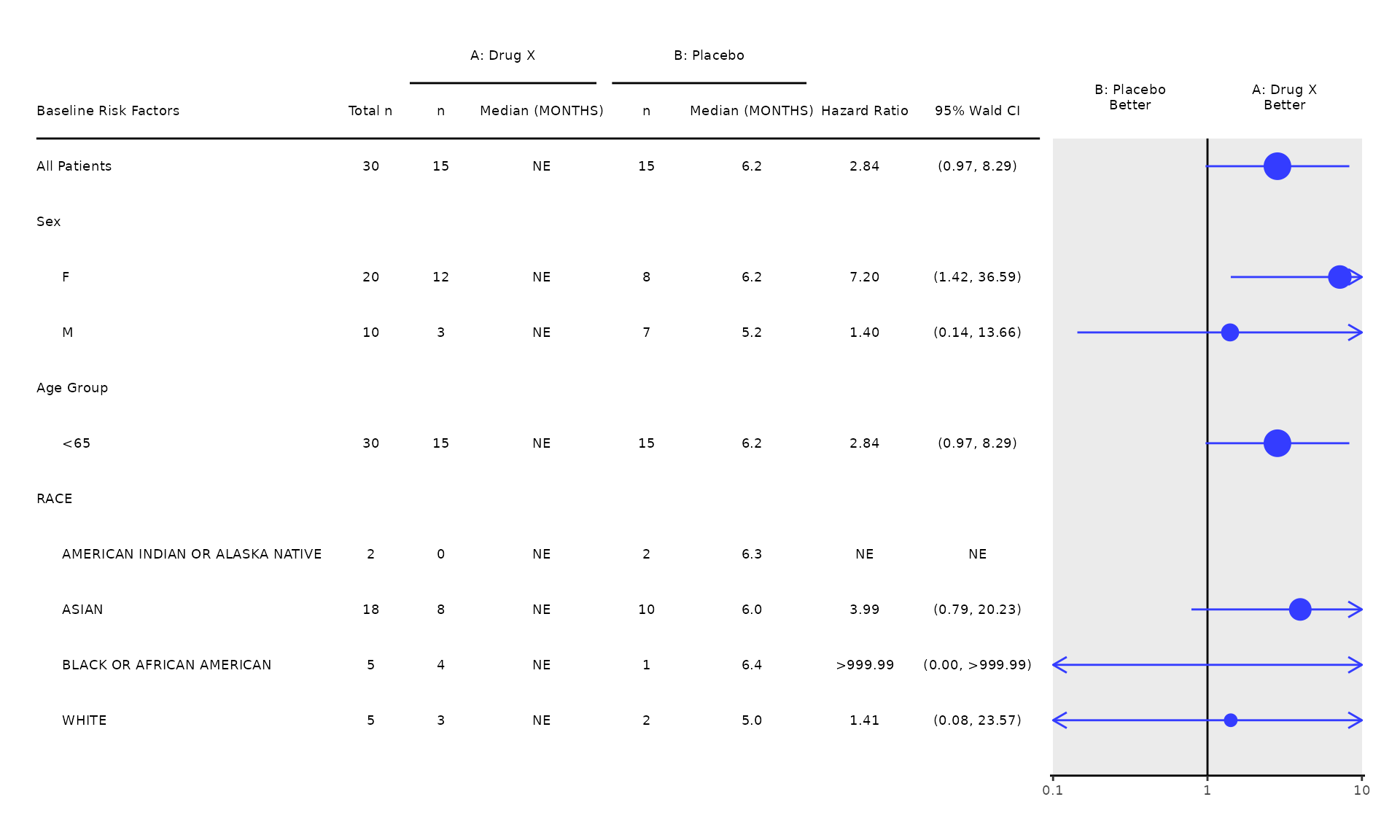
5. Forest Plot for Hazard Ratio (without proportional sizing of the hazard ratio symbol)
The col_symbol_size argument controls the size of the
hazard ratio symbols which are by default proportional in size to the
number of events in the subgroup. If NULL the same symbol
size is used for all subgroups.
run(fstg02, proc_data, col_symbol_size = NULL)
Kaplan-Meier Plot (KMG01)
1. Kaplan-Meier Plot (without comparative statistics)
- The
kmg01template produces the standard Kaplan-Meier Plot. - Users are expected to select a particular parameter for analysis.
- Users are expected to select the treatment groups to compare, otherwise, all treatment groups available in the input datasets will be plotted.
- The comparative statistics are not included by default.
- The estimation of median survival time per treatment group by default.
- More arguments in the
g_kmandcontrol_coxphfunctions can be passed through, please use the Help to find out more information.
proc_data <- log_filter(syn_data, PARAMCD == "OS", "adtte")
run(kmg01, proc_data, dataset = "adtte")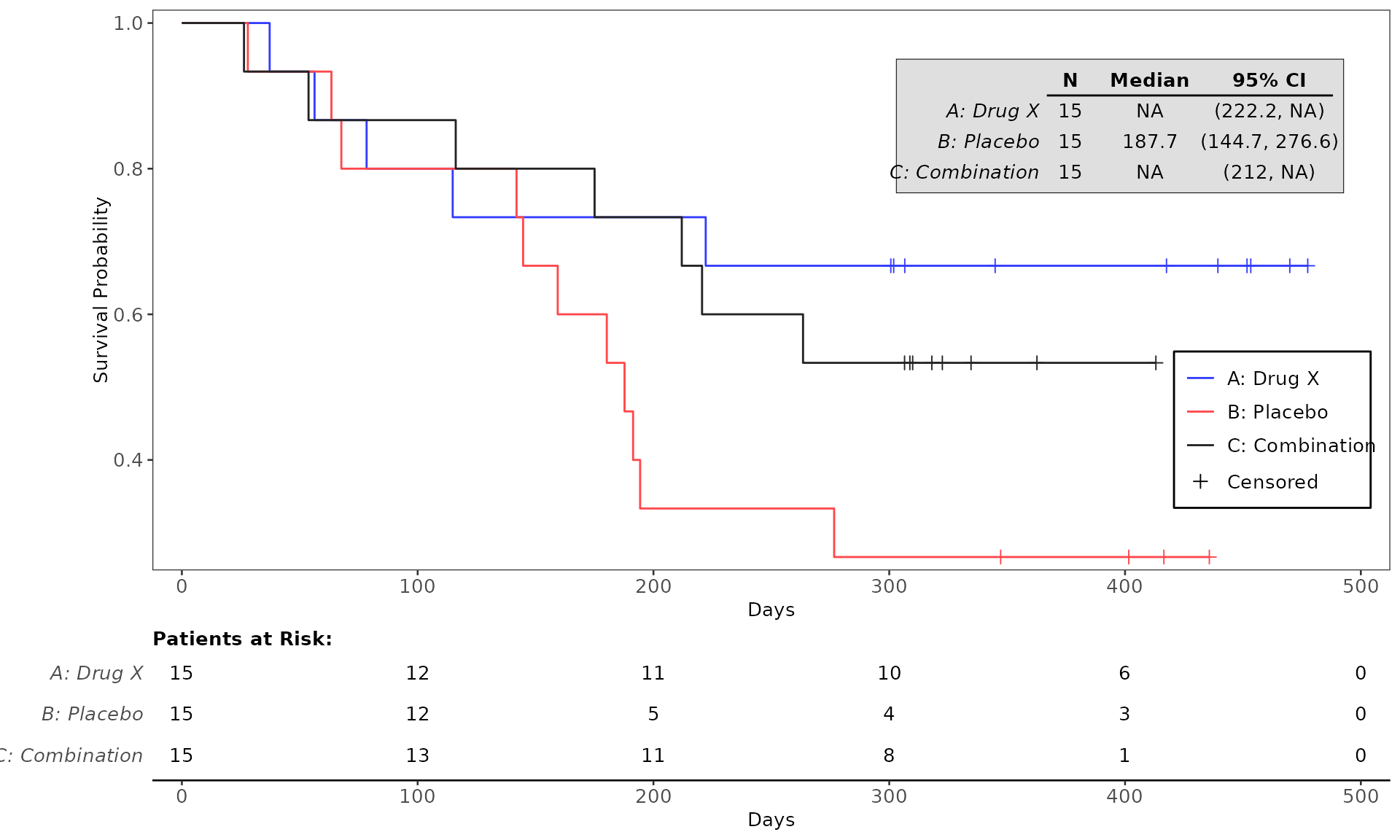
2. Kaplan-Meier Plot (with comparative statistics)
To enable the comparative statistics (hazard ratio and p-value), the
argument annot_coxph needs to be set to TRUE. The compare
group is determined by the levels in the factorized variable of
treatment group and the first level is used as reference group in the
statistics.
proc_data <- log_filter(syn_data, PARAMCD == "OS", "adtte")
run(
kmg01,
proc_data,
dataset = "adtte",
annot_coxph = TRUE,
control_annot_coxph = tern::control_coxph_annot(x = 0.33, y = 0.42)
)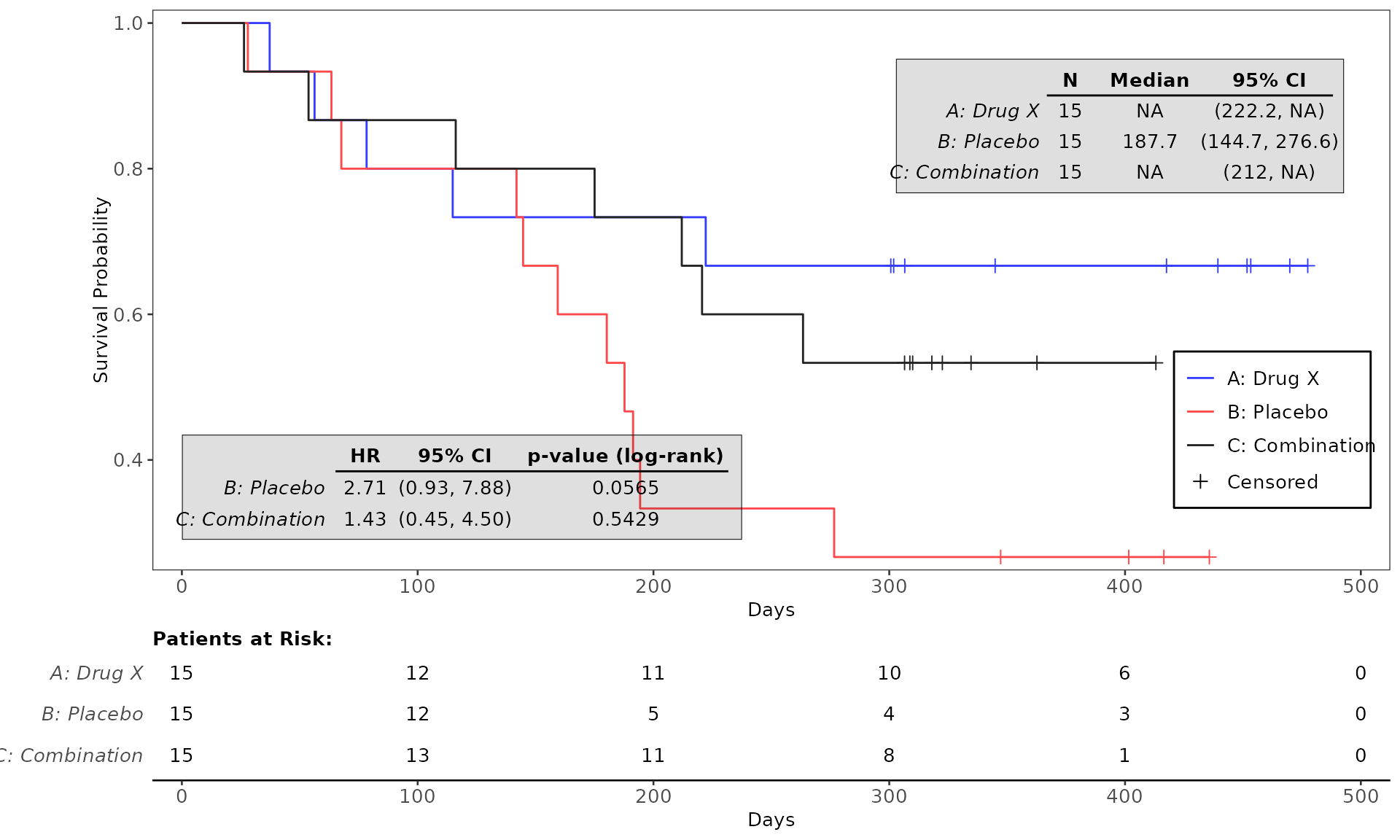
3. Kaplan-Meier Plot (without censoring marks)
To suppress the censoring marks, set the argument
cencor_show to FALSE.
proc_data <- log_filter(syn_data, PARAMCD == "OS", "adtte")
run(kmg01, proc_data, dataset = "adtte", censor_show = FALSE)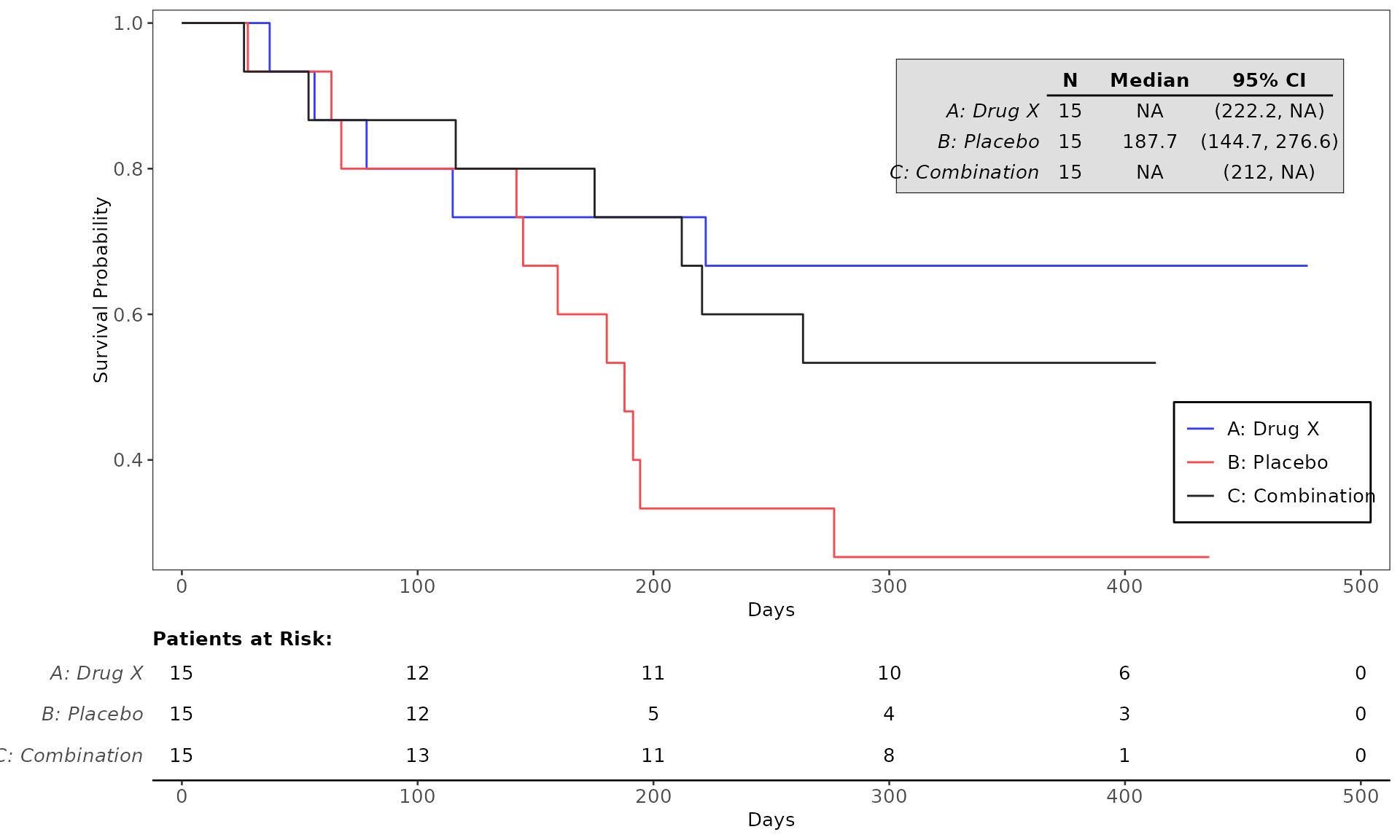
4. Kaplan-Meier Plot (without estimation of median survival time)
proc_data <- log_filter(syn_data, PARAMCD == "OS", "adtte")
run(kmg01, proc_data, dataset = "adtte", annot_surv_med = FALSE)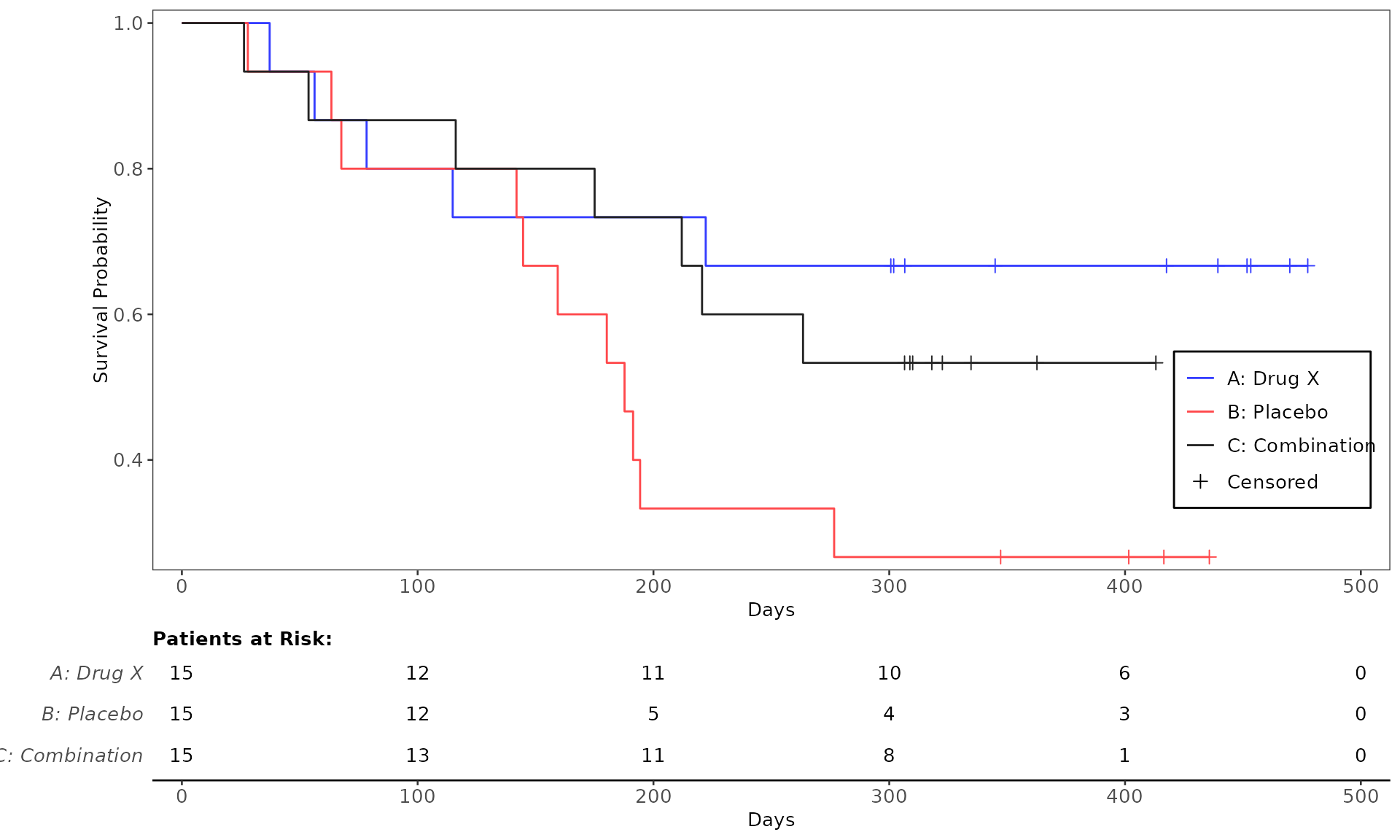
5. Kaplan-Meier Plot (with statistical annotation of either median or min of survival time)
To add the statistics annotation, use the function
annot_stats. Options are min or
median.
proc_data <- log_filter(syn_data, PARAMCD == "OS", "adtte")
run(kmg01, proc_data, dataset = "adtte", annot_stats = "median")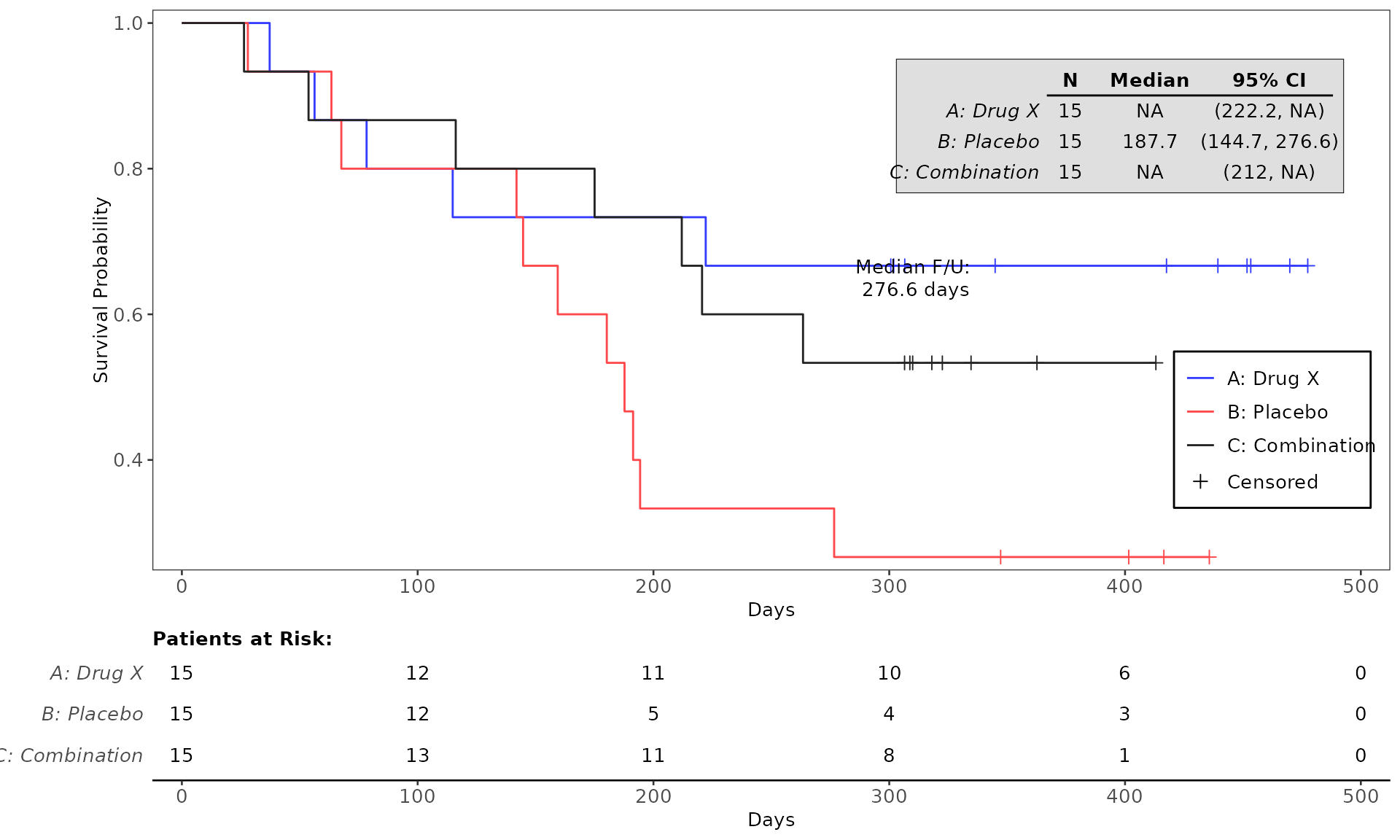
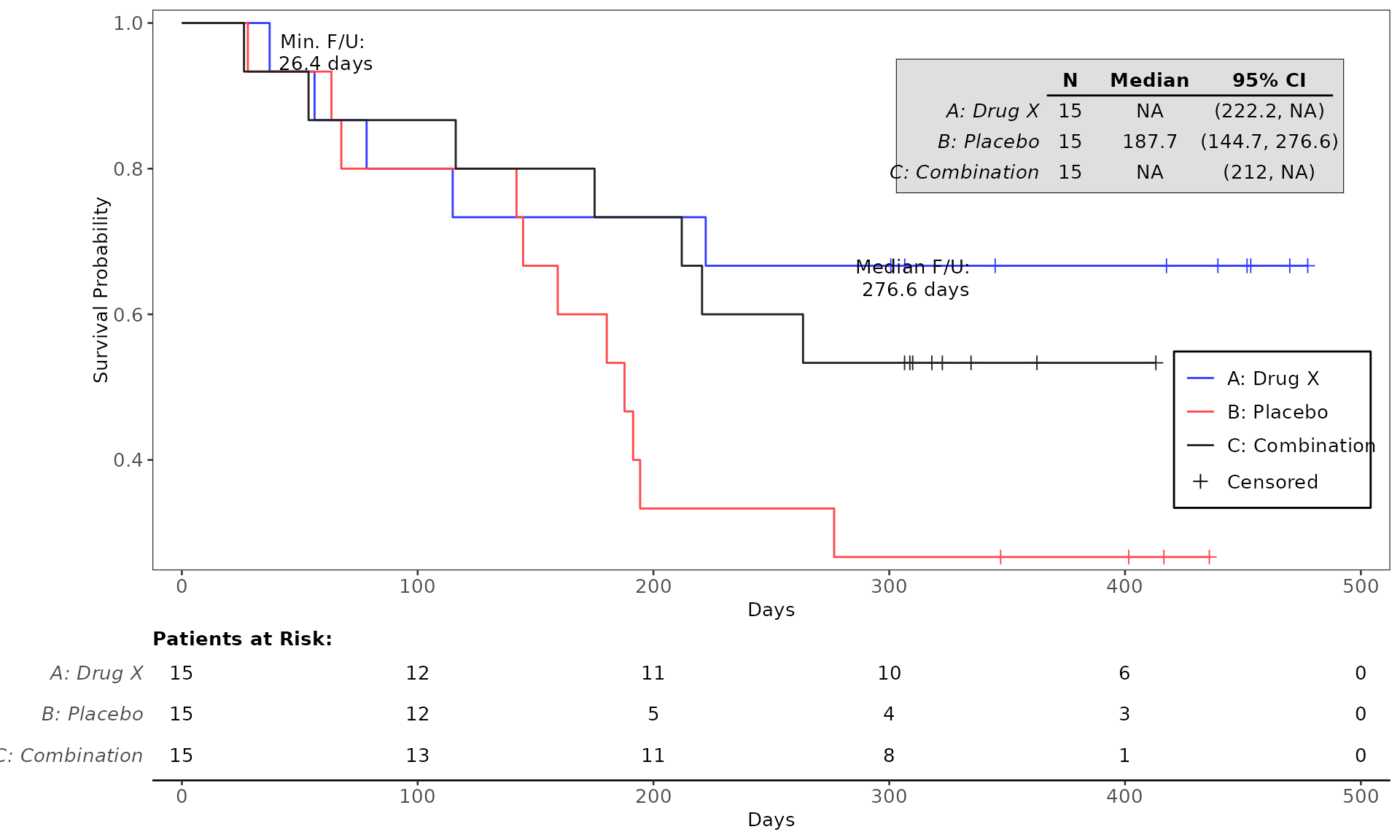
6. Kaplan-Meier Plot (without the table of patients at risk)
proc_data <- log_filter(syn_data, PARAMCD == "OS", "adtte")
run(kmg01, proc_data, dataset = "adtte", annot_at_risk = FALSE)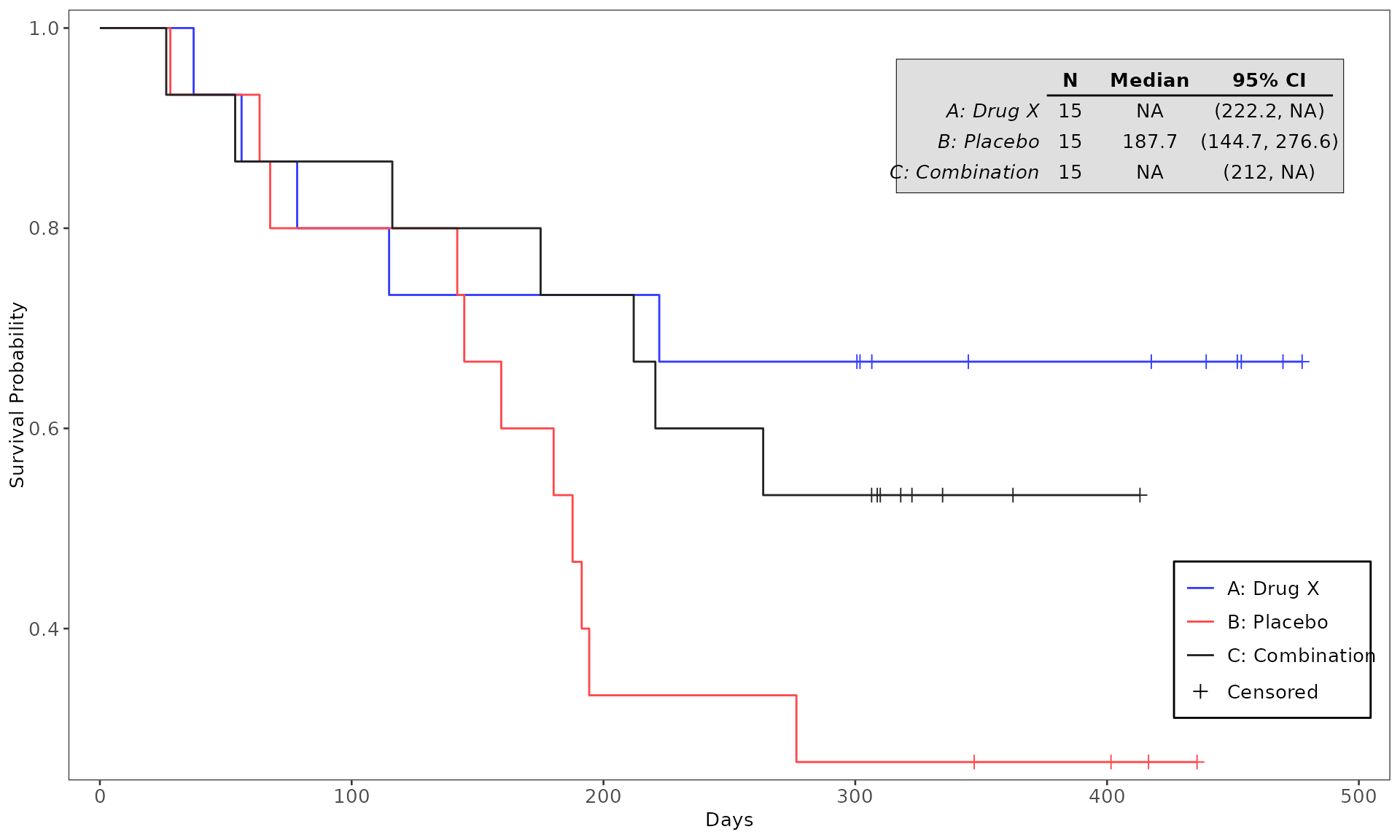
Mean Plot (MNG01)
1. Plot of Mean and Confidence Interval (with Table Section)
- The
mng01template produces the standard mean plot. - Note that the template
mng01is quite general. The users are expected to specify the analysis dataset and the visit variable in therunfunction, and select the parameters prior to therunfunction. - The table of summary statistics is included by default.
- The variable Analysis Value
AVALis used for plotting by default. - If the input dataset contains results of the same analyses in
multiple units,(e.g. SI/CV units in
ADLB), please make sure that the parameters in appropriate units are selected in advance.
proc_data <- log_filter(syn_data, PARAMCD == "DIABP", "advs")
run(mng01, proc_data, dataset = "advs", x_var = c("AVISIT", "AVISITN"))
#> $`Diastolic Blood Pressure`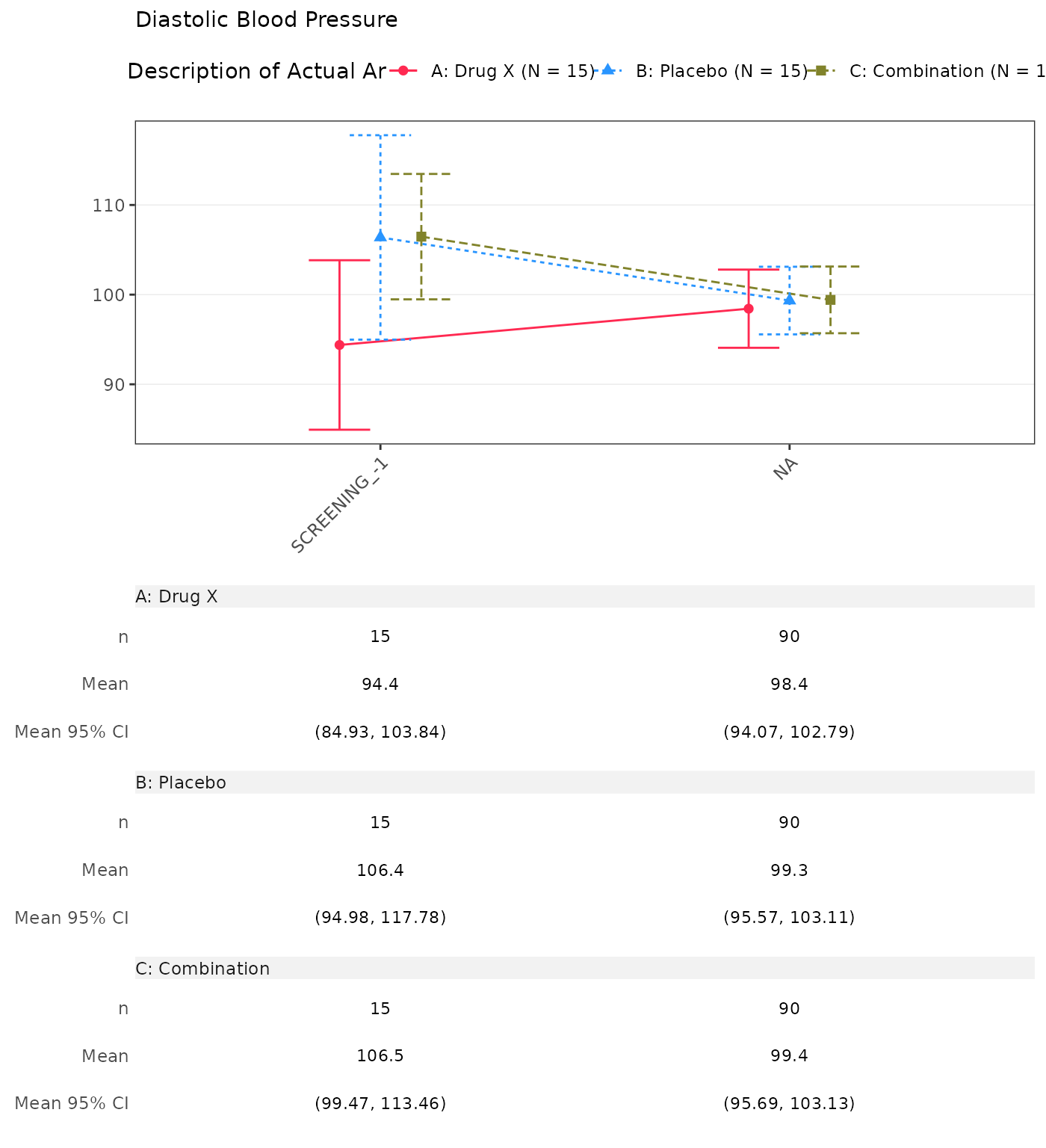
2. Plot of Mean and Confidence Interval of Change from Baseline of Vital Signs
proc_data <- log_filter(syn_data, PARAMCD == "DIABP", "advs")
run(mng01, proc_data, dataset = "advs", x_var = c("AVISIT", "AVISITN"), y_var = "CHG")
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> $`Diastolic Blood Pressure`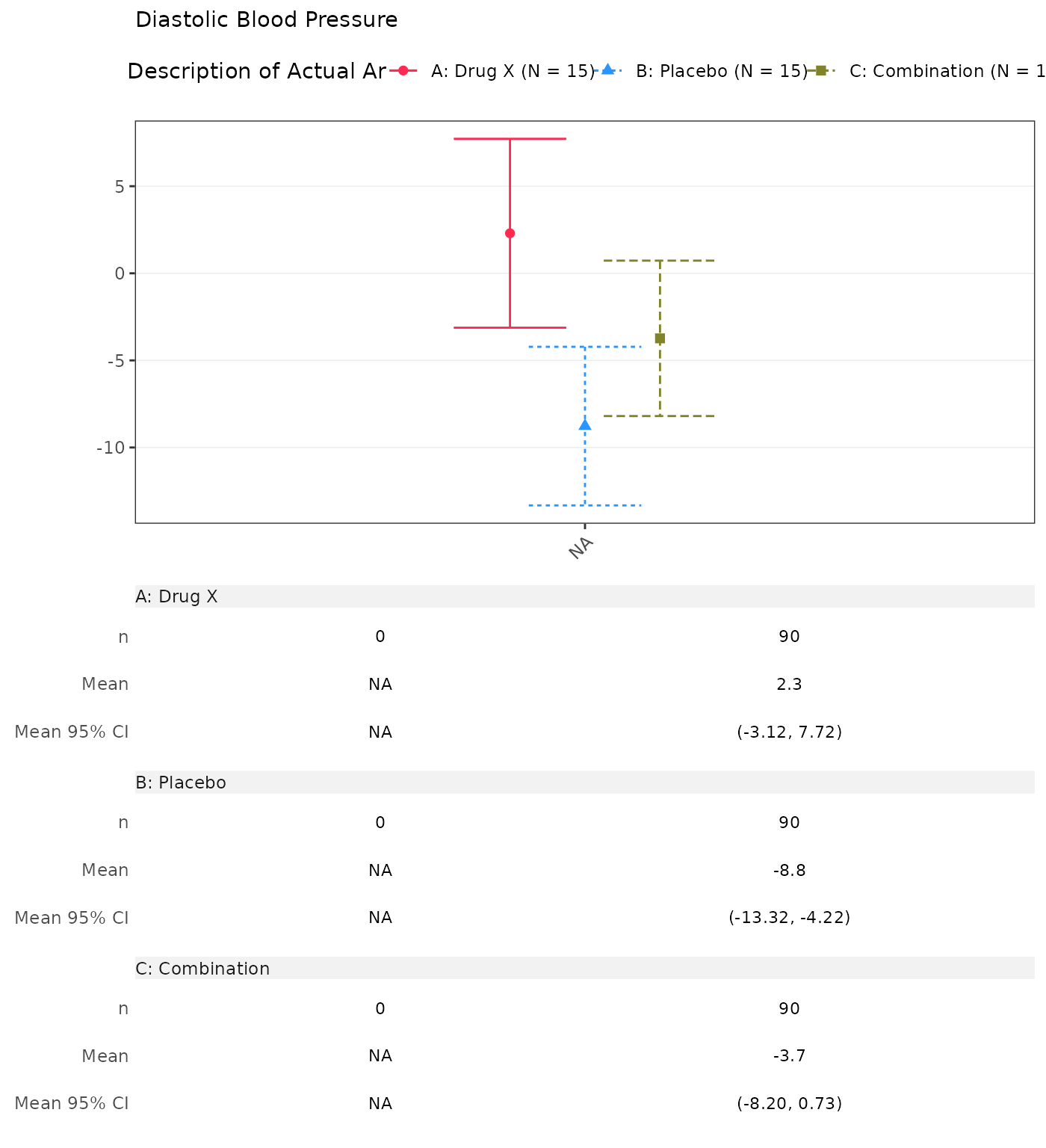
3. Plot of Mean (+/-SD) (Changing the Statistics)
To change the statistics, use the argument interval_fun.
Options are mean_ci, mean_sei,
mean_sdi, median_ci,
quantiles,range.
proc_data <- log_filter(syn_data, PARAMCD == "DIABP", "advs")
run(mng01, proc_data, dataset = "advs", x_var = c("AVISIT", "AVISITN"), interval_fun = "mean_sdi")
#> $`Diastolic Blood Pressure`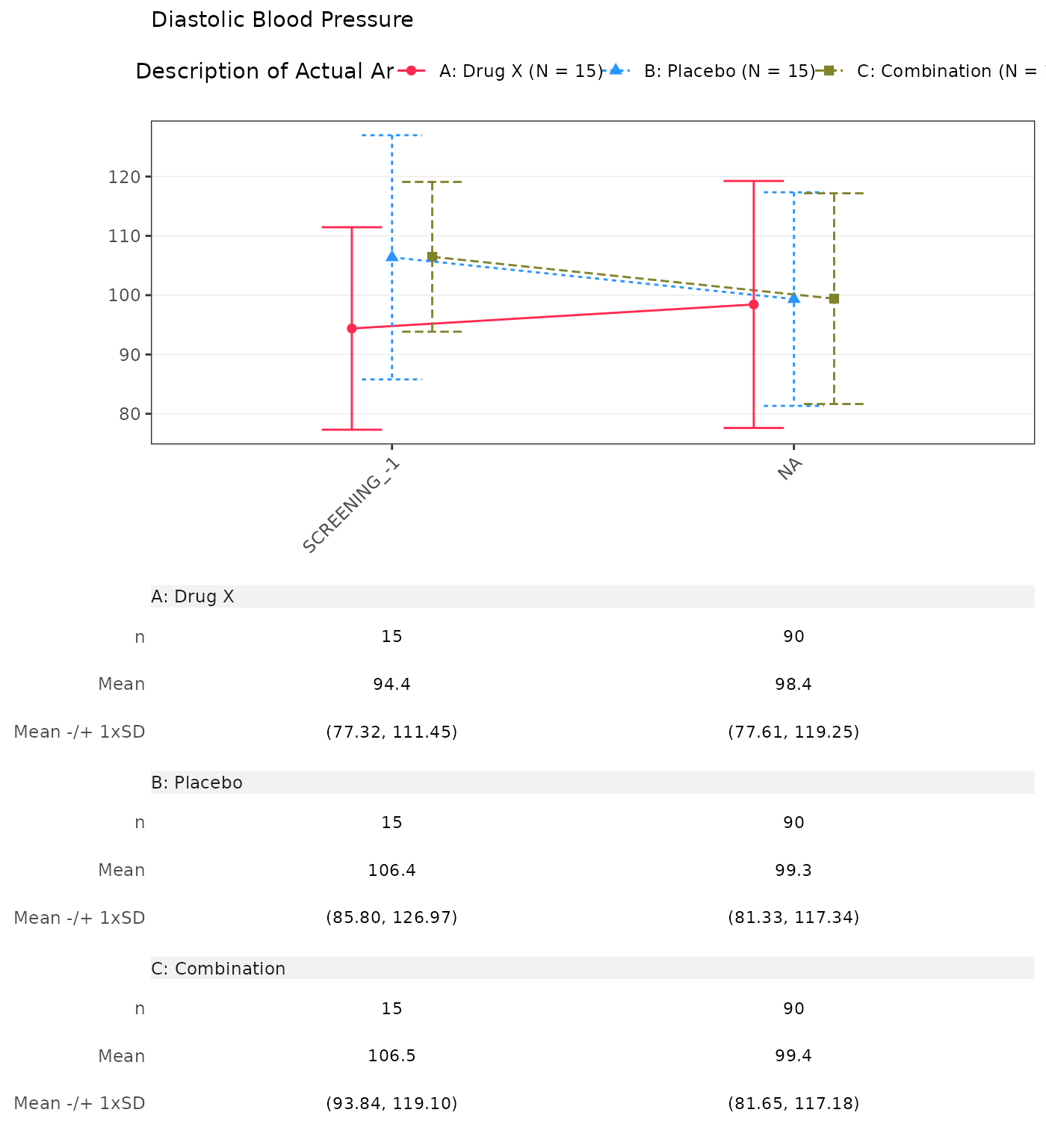
4. Plot of Mean and Confidence Interval (Modify Alpha Level)
To change the alpha level of the confidence interval, use the
argument
control = control_analyze_vars(conf_level = <0.xx>).
Note that this is only in effect when interval_fun is set
to mean_ci.
proc_data <- log_filter(syn_data, PARAMCD == "DIABP", "advs")
run(
mng01, proc_data,
dataset = "advs", x_var = c("AVISIT", "AVISITN"),
interval_fun = "mean_ci", control = tern::control_analyze_vars(conf_level = 0.80)
)
#> $`Diastolic Blood Pressure`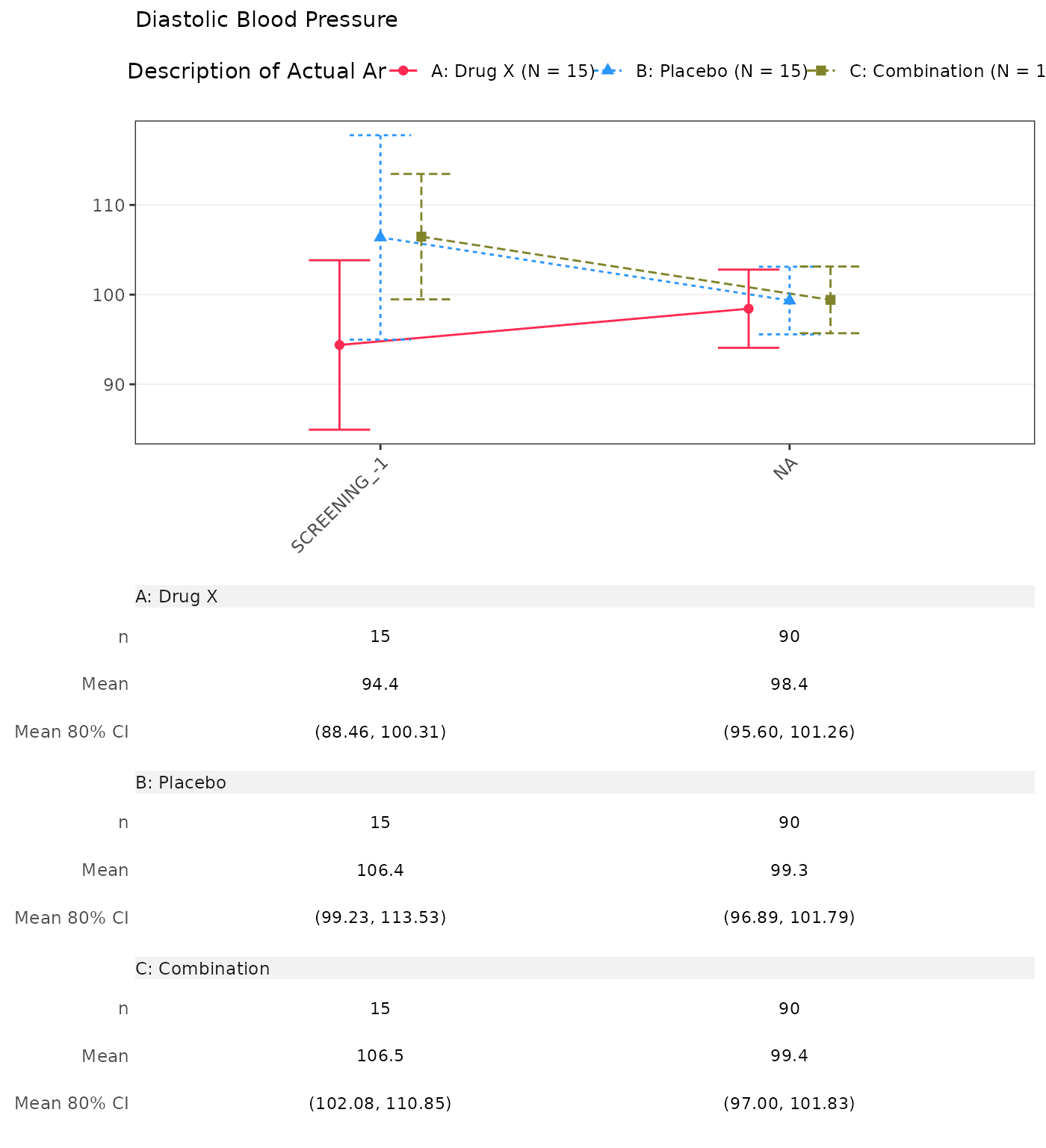
5. Plot of Mean and Confidence Interval (With Number of Patients Only)
proc_data <- log_filter(syn_data, PARAMCD == "DIABP", "advs")
run(mng01, proc_data, dataset = "advs", x_var = c("AVISIT", "AVISITN"), table = "n")
#> $`Diastolic Blood Pressure`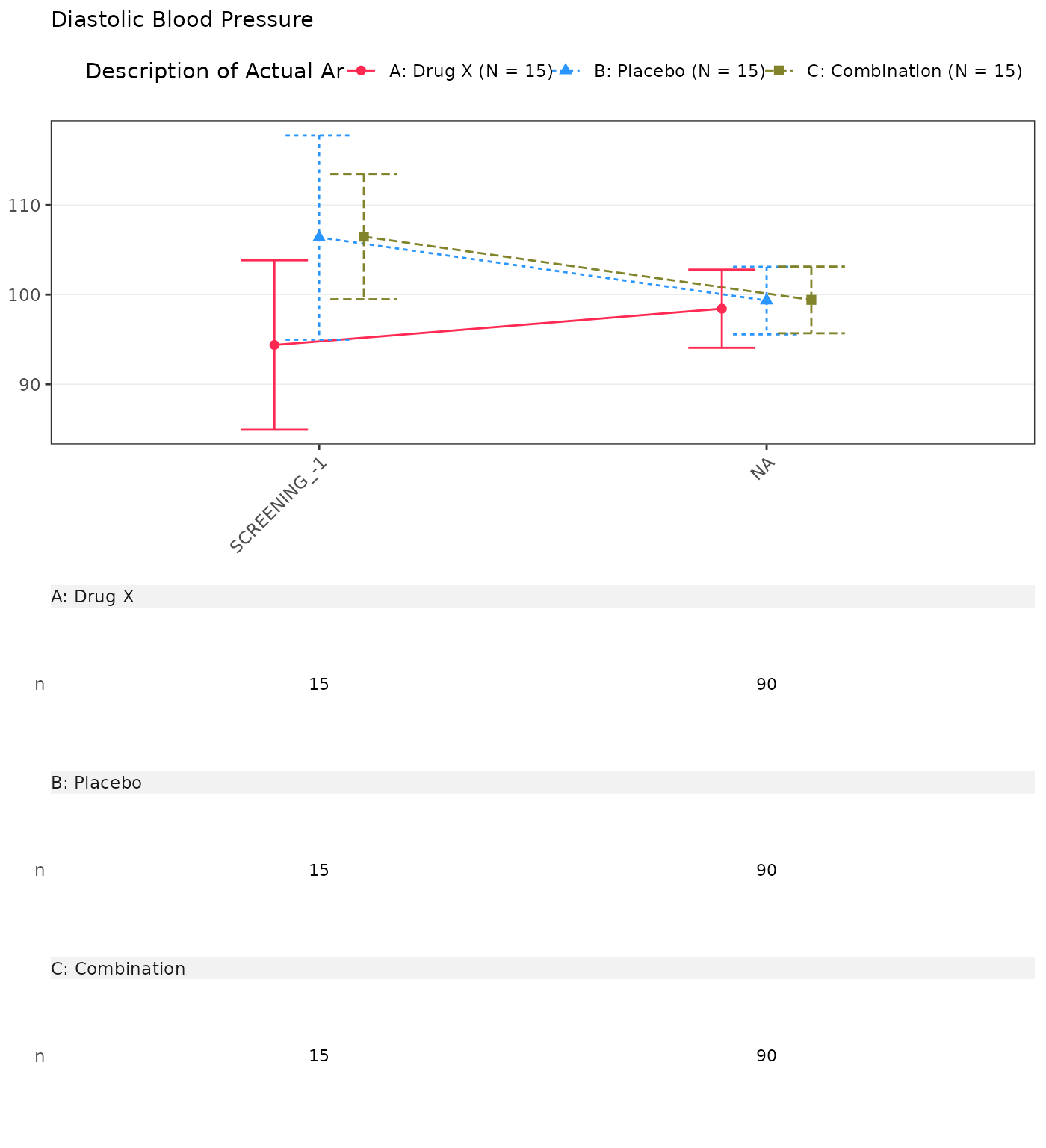
6. Plot of Mean and Confidence Interval (without Table Section)
proc_data <- log_filter(syn_data, PARAMCD == "DIABP", "advs")
run(mng01, proc_data, dataset = "advs", x_var = c("AVISIT", "AVISITN"), table = NULL)
#> $`Diastolic Blood Pressure`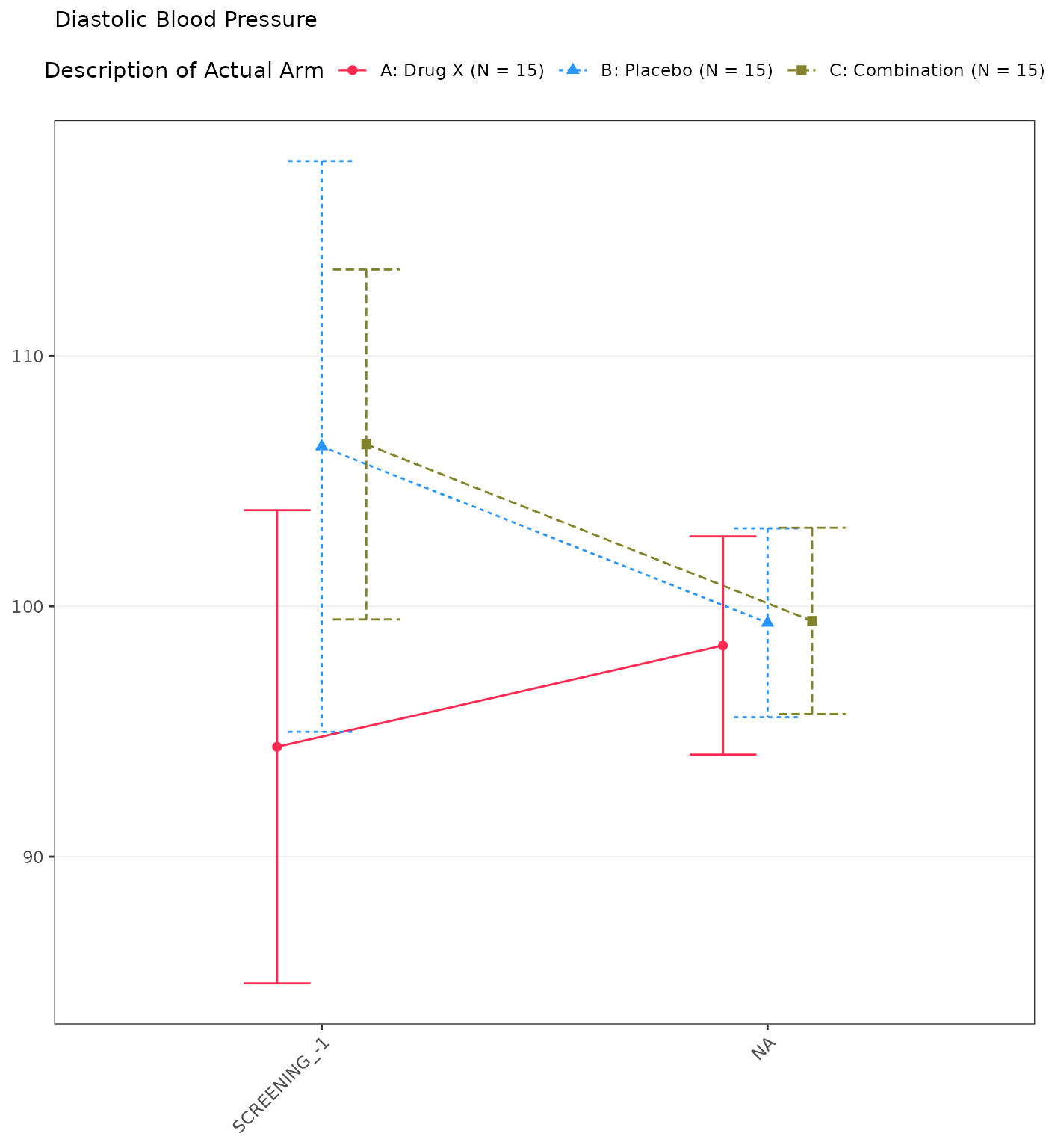
A new argument has been added to control the theme (e.g. setting the angle of the axis); see an example below:
ggtheme <- ggplot2::theme(
panel.grid = ggplot2::element_line(colour = "black", linetype = 3),
panel.background = ggplot2::element_rect(fill = "white"),
legend.position = "top",
axis.text.x = ggplot2::element_text(angle = 22, hjust = 1, vjust = 1)
)
run(mng01, syn_data, dataset = "adlb", ggtheme = ggtheme)
#> $`Alanine Aminotransferase Measurement`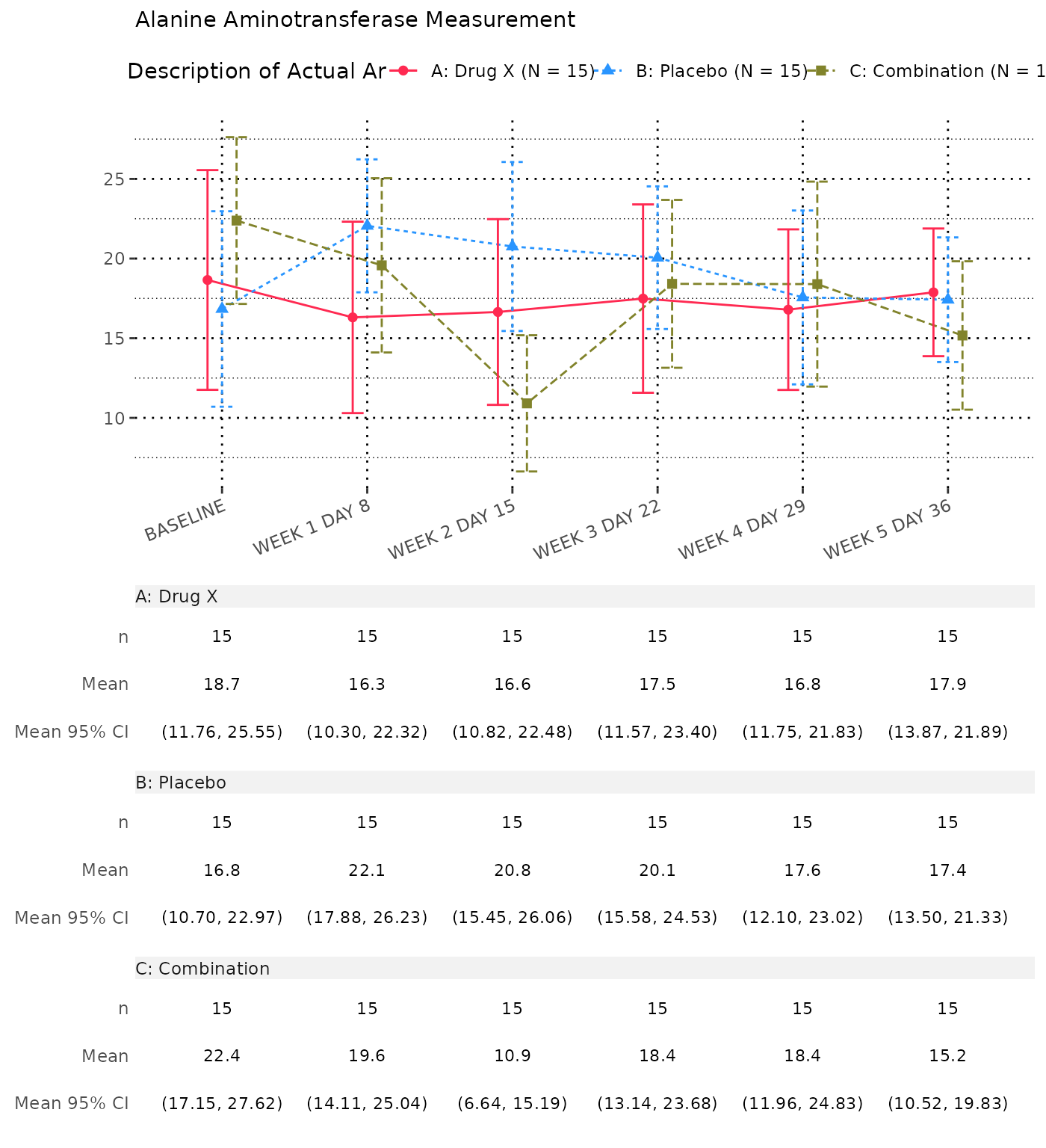
#>
#> $`C-Reactive Protein Measurement`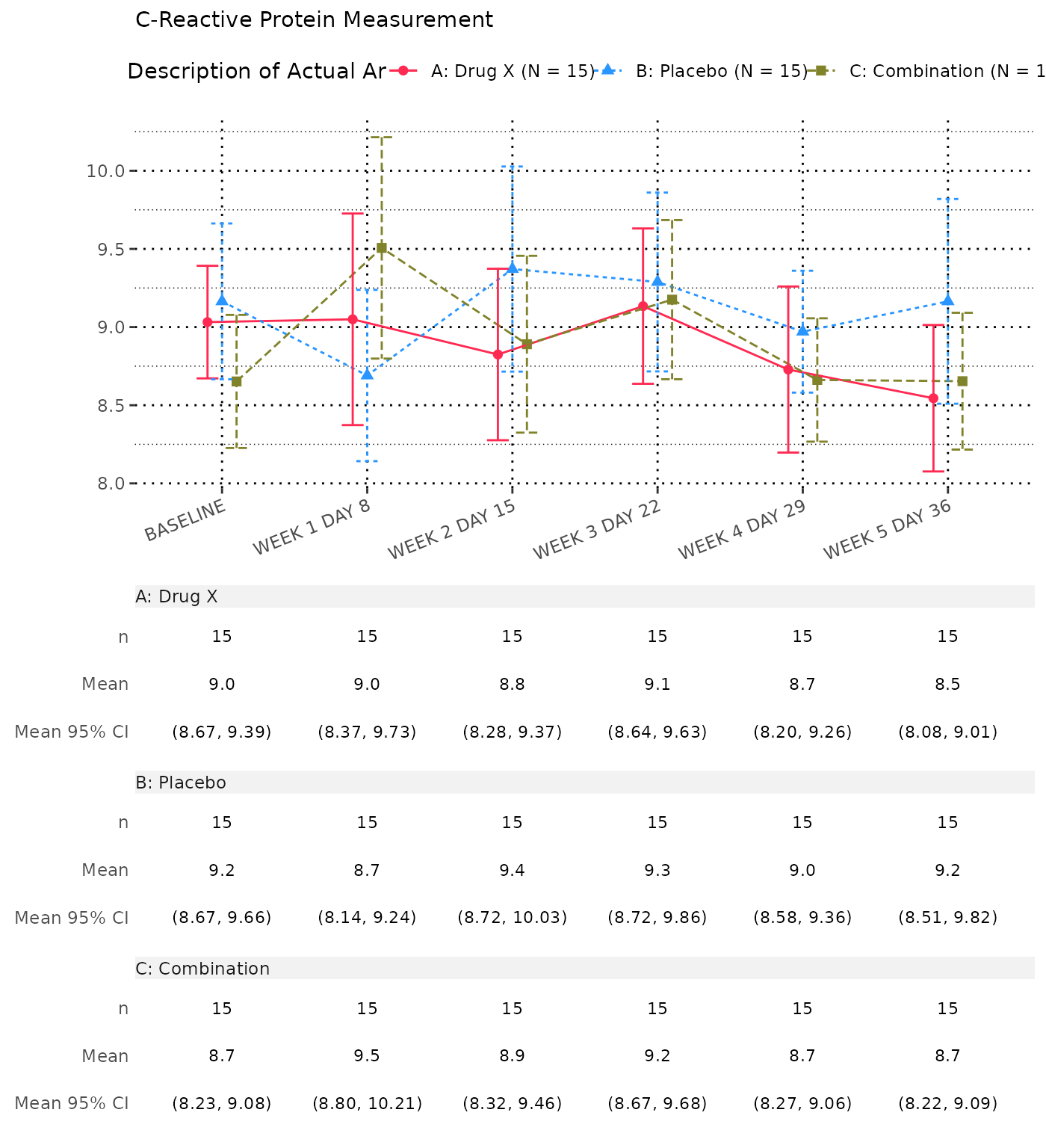
#>
#> $`Immunoglobulin A Measurement`