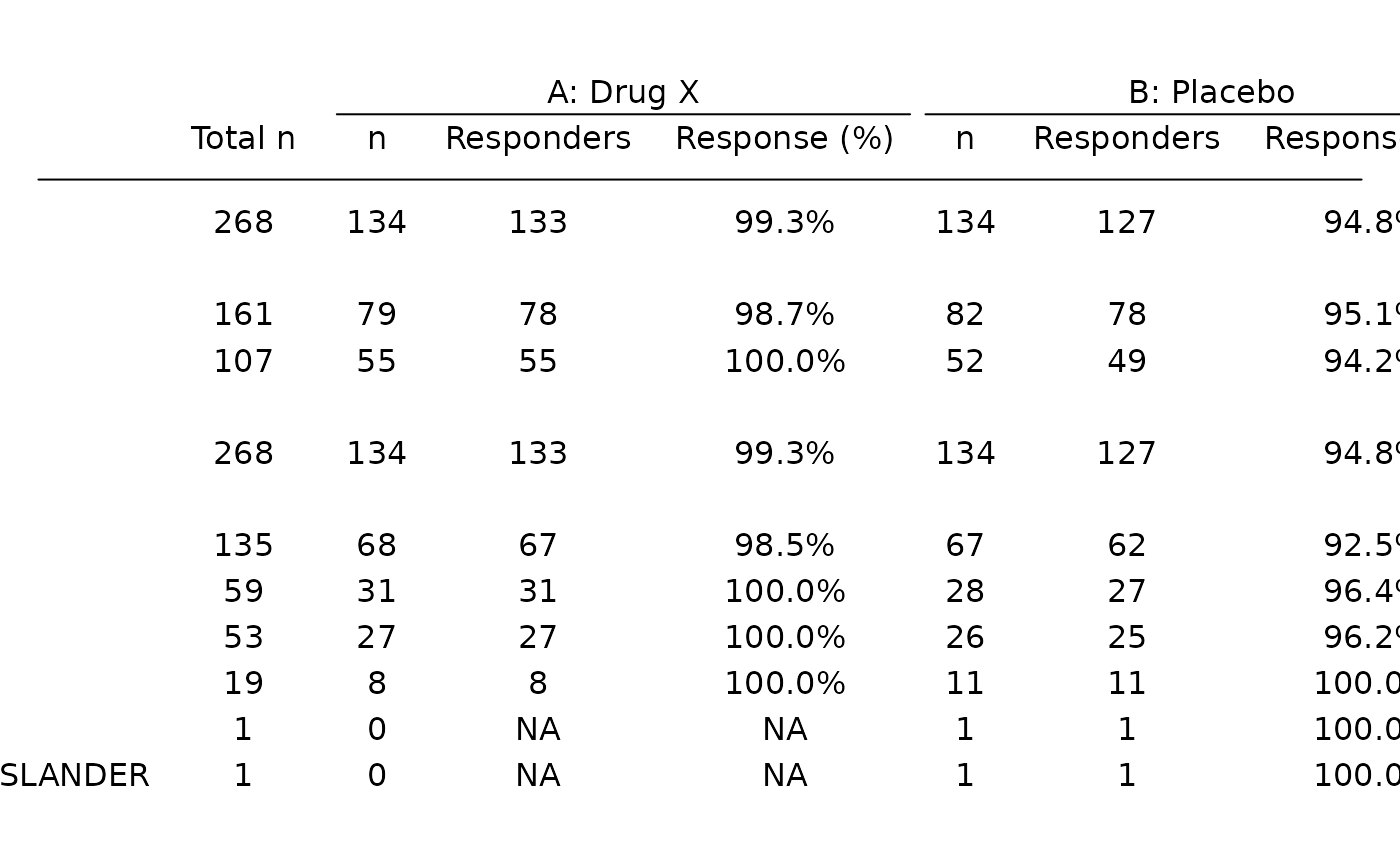
FSTG01 Subgroup Analysis of Best Overall Response.
fstg01.RdFSTG01 Subgroup Analysis of Best Overall Response.
Usage
fstg01_main(
adam_db,
dataset = "adrs",
arm_var = "ARM",
rsp_var = "is_rsp",
subgroups = c("SEX", "AGEGR1", "RACE"),
strata_var = NULL,
...
)
fstg01_pre(adam_db, ...)
fstg01_post(tlg, ...)
fstg01Arguments
- adam_db
(
listofdata.frames) object containing theADaMdatasets- dataset
(
string) the name of a table in theadam_dbobject.- arm_var
(
string) the arm variable name used for group splitting.- rsp_var
(
string) the response variable name to flag whether each subject is a binary response or not.- subgroups
(
character) the subgroups variable name to list baseline risk factors.- strata_var
(
character) required if stratified analysis is performed.- ...
Further arguments passed to
g_forestandextract_rsp_subgroups(a wrapper forh_odds_ratio_subgroups_dfandh_proportion_subgroups_df). For details, see the documentation intern. Commonly used arguments includecol_symbol_size,col,vline,groups_lists,conf_level,method,label_all, etc.- tlg
(
TableTree,Listingorggplot) object typically produced by amainfunction.
Functions
fstg01_main(): Main TLG Functionfstg01_pre(): Preprocessingfstg01_post(): Postprocessing