KMG01 Kaplan-Meier Plot 1.
kmg01.RdKMG01 Kaplan-Meier Plot 1.
Usage
kmg01_main(adam_db, dataset = "adtte", arm_var = "ARM", ...)
kmg01_pre(adam_db, dataset = "adtte", ...)
kmg01_post(tlg, ...)
kmg01Arguments
- adam_db
(
listofdata.frames) object containing theADaMdatasets- dataset
(
string) the name of a table in theadam_dbobject.- arm_var
(
string) variable used for column splitting- ...
Further arguments passed to
g_kmandcontrol_coxph. For details, see the documentation intern. Commonly used arguments includecol,pval_method,ties,conf_level,conf_type,annot_coxph,annot_stats, etc.- tlg
(
TableTree,Listingorggplot) object typically produced by amainfunction.
Note
adam_dbobject must contain the table specified bydatasetwith the columns specified byarm_var.
Examples
library(dplyr)
library(dunlin)
col <- c(
"A: Drug X" = "black",
"B: Placebo" = "blue",
"C: Combination" = "gray"
)
syn_data2 <- log_filter(syn_data, PARAMCD == "OS", "adtte")
run(kmg01, syn_data2, dataset = "adtte", line_col = col)
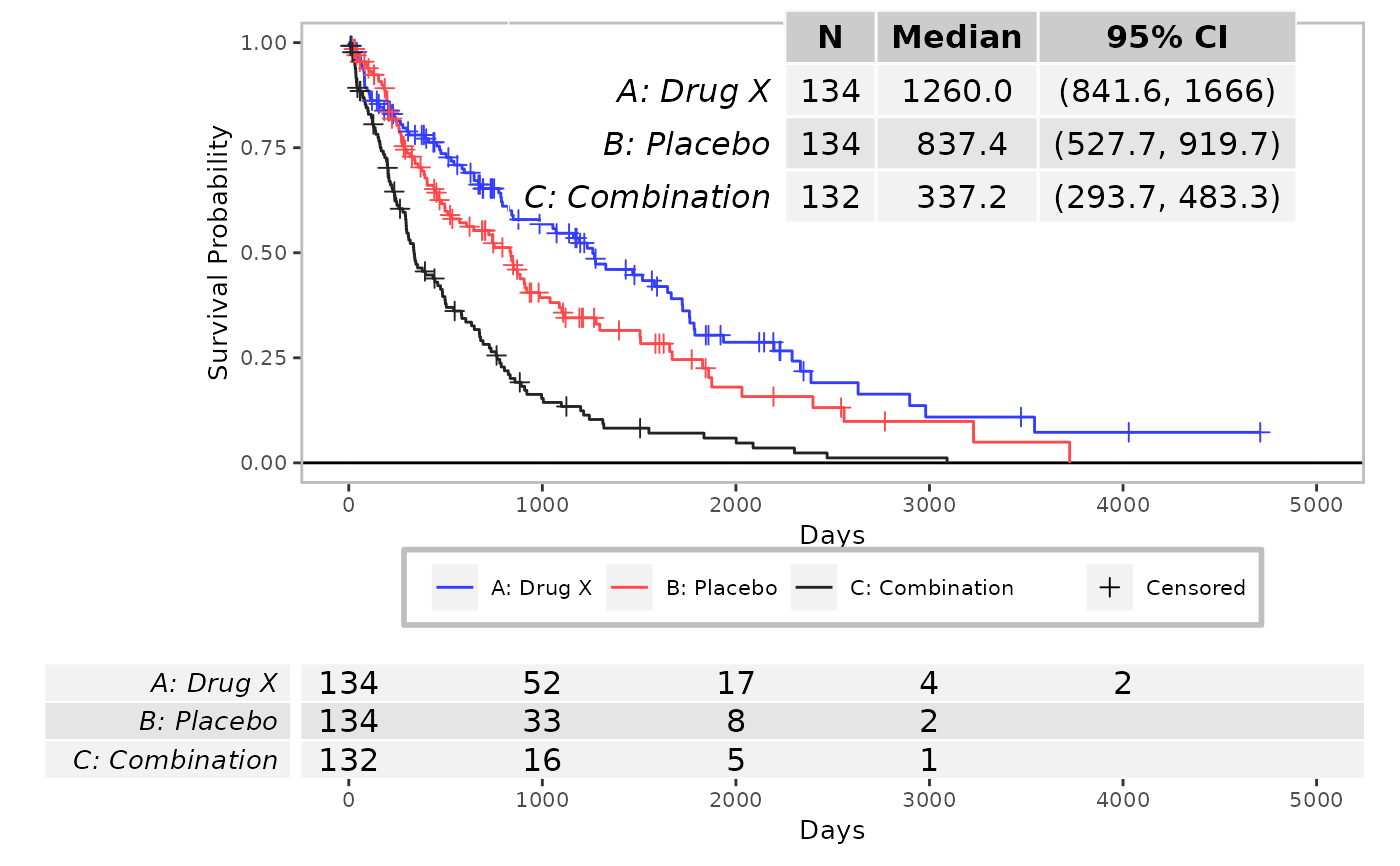 #> gTree[GRID.gTree.216]
syn_data3 <- log_filter(syn_data, PARAMCD == "AEREPTTE", "adaette")
run(kmg01, syn_data3, dataset = "adaette")
#> gTree[GRID.gTree.216]
syn_data3 <- log_filter(syn_data, PARAMCD == "AEREPTTE", "adaette")
run(kmg01, syn_data3, dataset = "adaette")
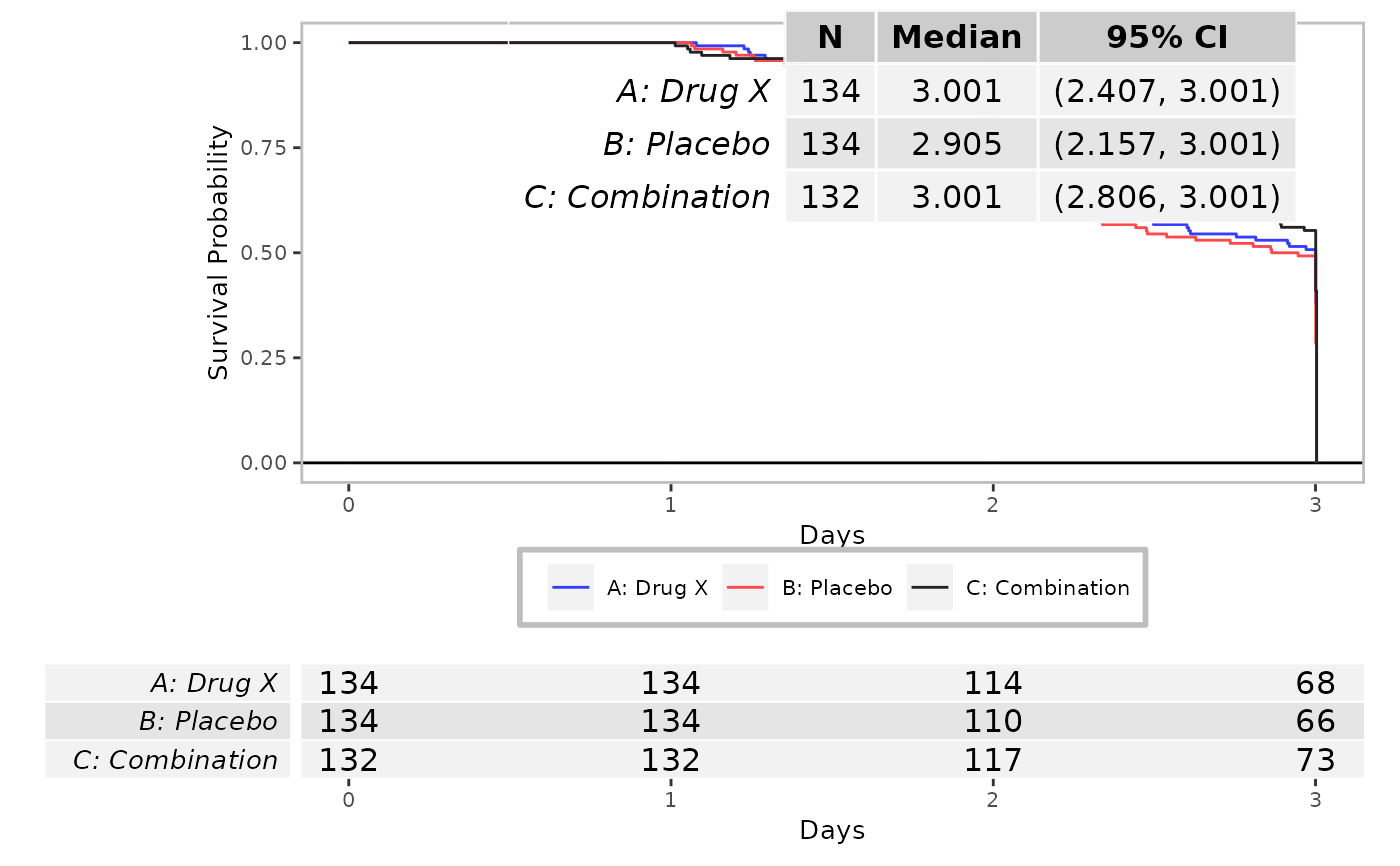 #> gTree[GRID.gTree.329]
#> gTree[GRID.gTree.329]