This produces boxplots of the gene expression values of a single gene, multiple genes or a gene signature.
Usage
draw_boxplot(
object,
assay_name,
genes,
x_var = NULL,
color_var = NULL,
facet_var = NULL,
violin = FALSE,
jitter = FALSE
)
h_draw_boxplot_df(object, assay_name, genes, x_var, color_var, facet_var)Arguments
- object
(
AnyHermesData)
input.- assay_name
(
string)
selects assay from input for the y-axis.- genes
(
GeneSpec)
for which genes or which gene signature to produce boxplots.- x_var
(
stringorNULL)
optional stratifying variable for the x-axis, taken from input sample variables.- color_var
(
stringorNULL)
optional color variable, taken from input sample variables.- facet_var
(
stringorNULL)
optional faceting variable, taken from input sample variables.- violin
(
flag)
whether to draw a violin plot instead of a boxplot.- jitter
(
flag)
whether to add jittered original data points.
Examples
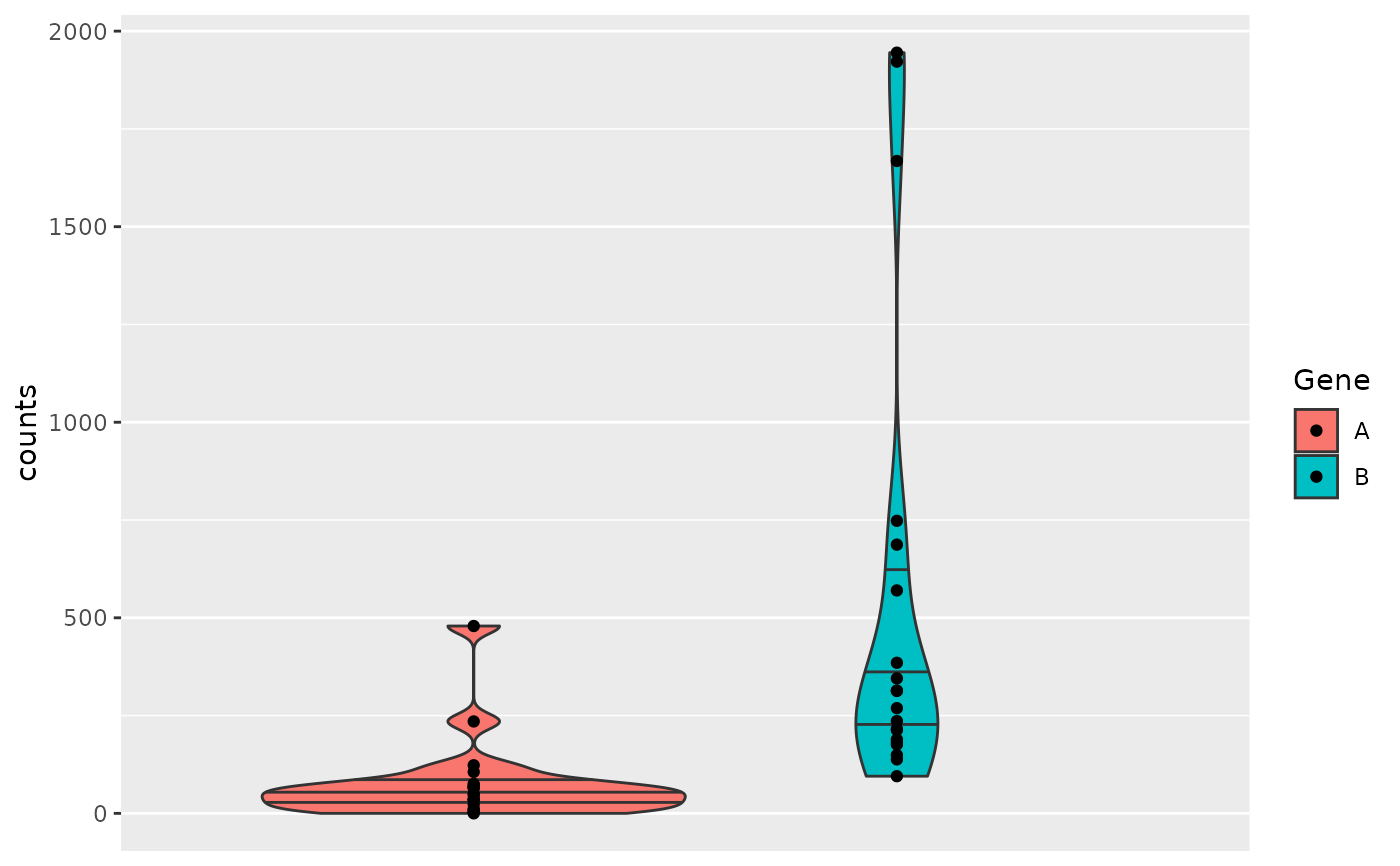
object <- hermes_data
draw_boxplot(
object,
assay_name = "counts",
genes = gene_spec(c(A = genes(object)[1])),
violin = TRUE
)
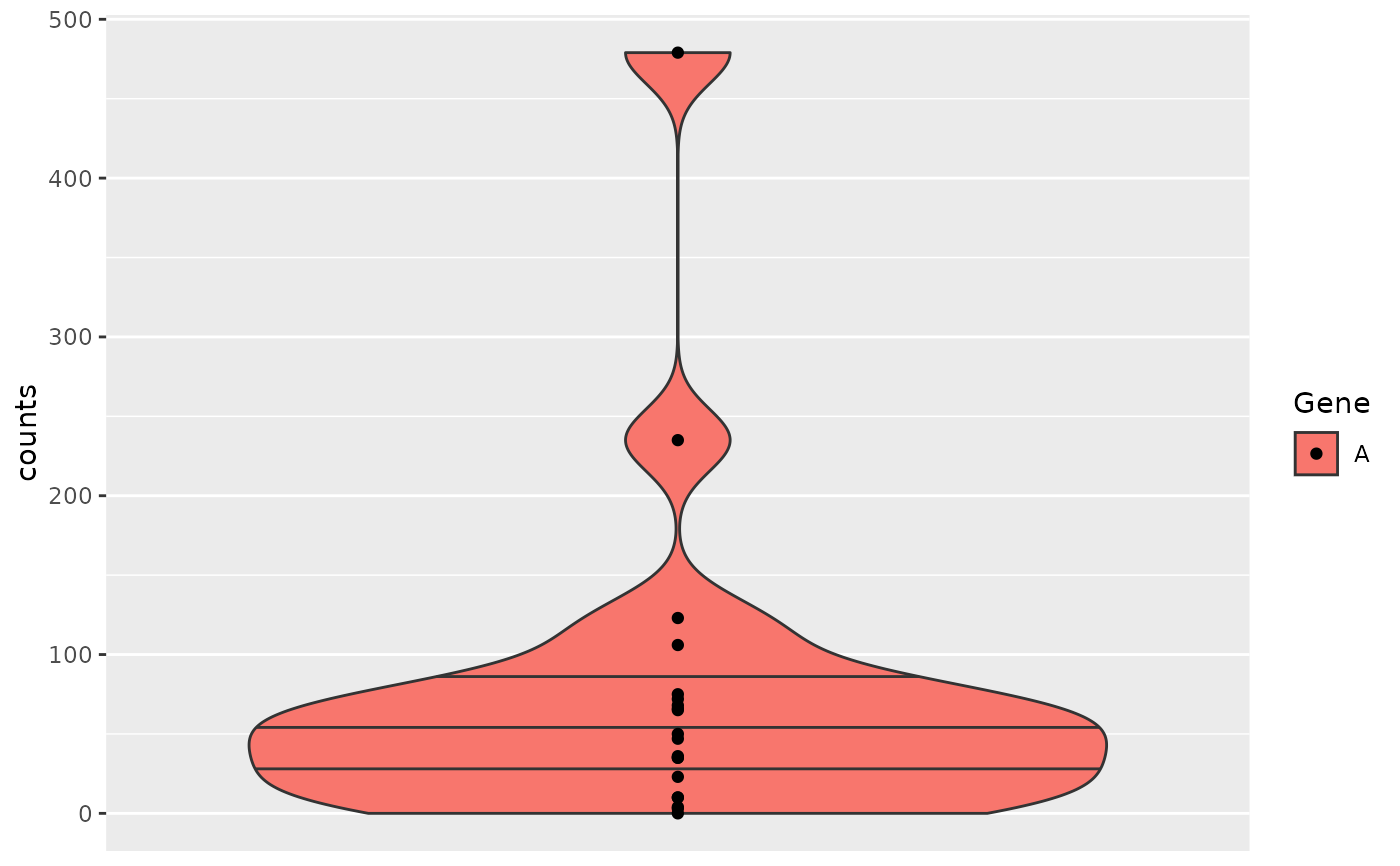 object2 <- object %>%
add_quality_flags() %>%
filter() %>%
normalize()
draw_boxplot(
object2,
assay_name = "tpm",
x_var = "SEX",
genes = gene_spec(setNames(genes(object2)[1:10], 1:10), fun = colMeans),
facet_var = "RACE",
color_var = "AGE18",
jitter = TRUE
)
object2 <- object %>%
add_quality_flags() %>%
filter() %>%
normalize()
draw_boxplot(
object2,
assay_name = "tpm",
x_var = "SEX",
genes = gene_spec(setNames(genes(object2)[1:10], 1:10), fun = colMeans),
facet_var = "RACE",
color_var = "AGE18",
jitter = TRUE
)
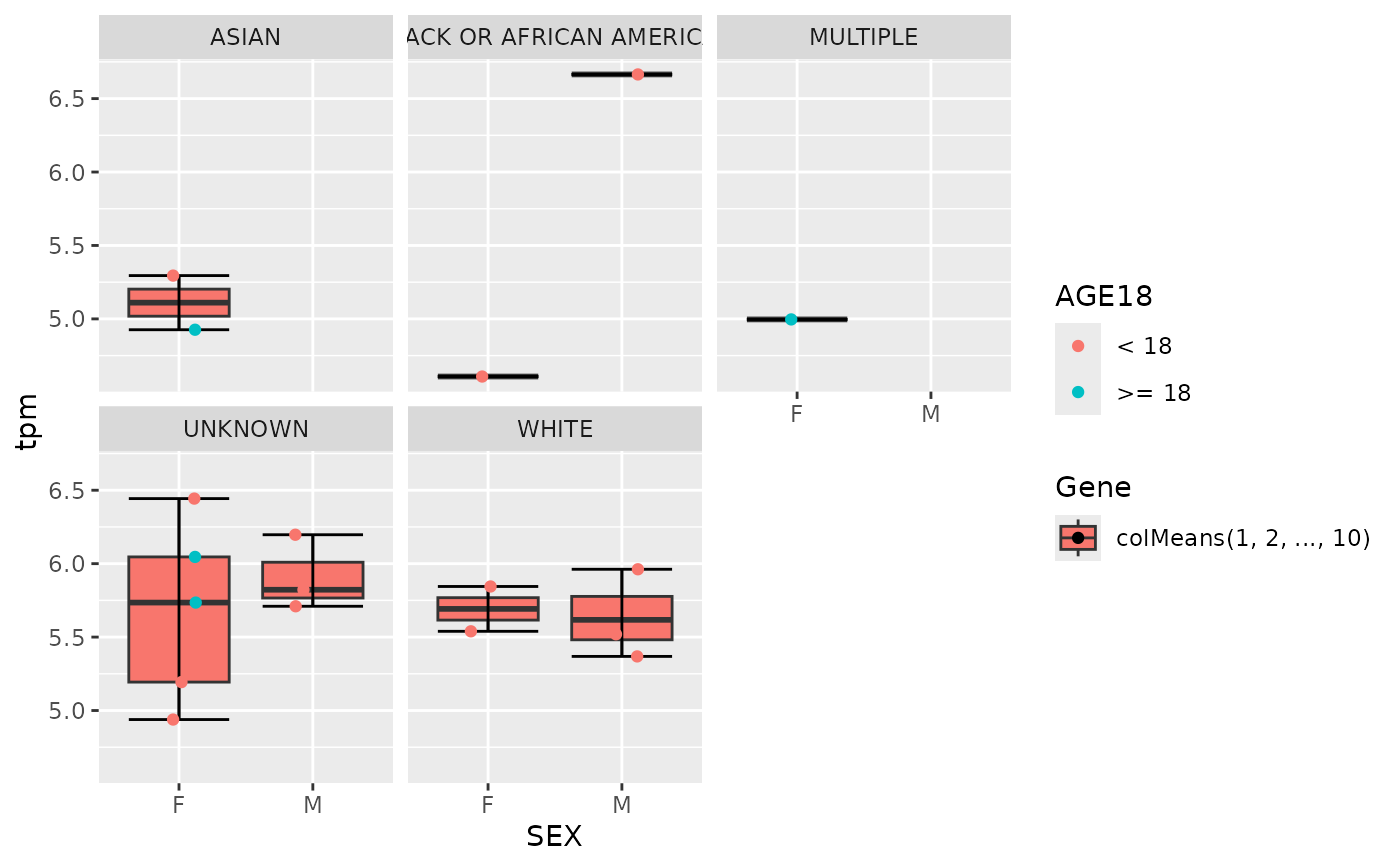 draw_boxplot(
object,
assay_name = "counts",
x_var = "SEX",
genes = gene_spec(genes(object)[1:3]),
jitter = TRUE,
facet_var = "AGE18"
)
draw_boxplot(
object,
assay_name = "counts",
x_var = "SEX",
genes = gene_spec(genes(object)[1:3]),
jitter = TRUE,
facet_var = "AGE18"
)
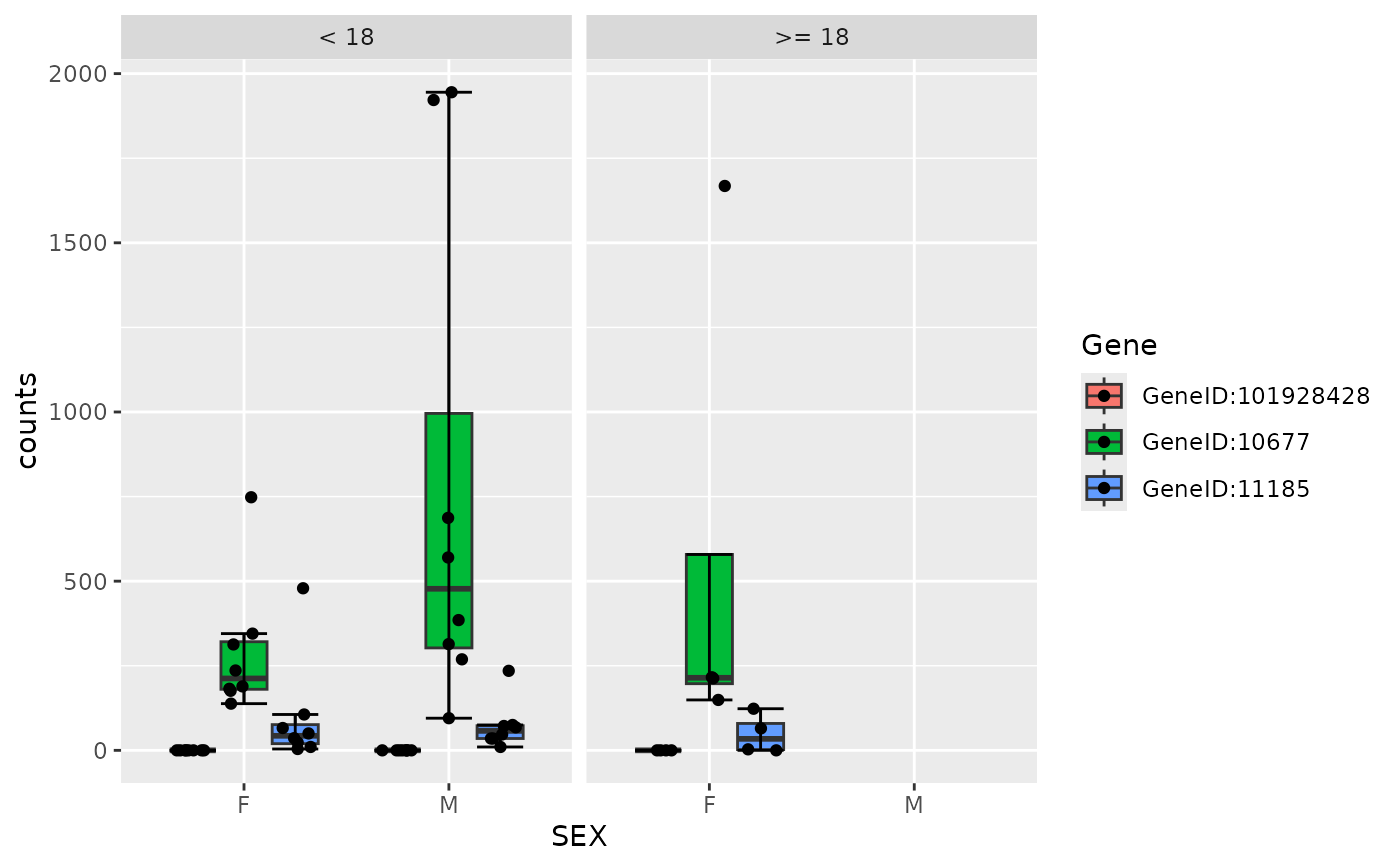 draw_boxplot(
object,
assay_name = "counts",
genes = gene_spec(c(A = "GeneID:11185", B = "GeneID:10677")),
violin = TRUE
)
draw_boxplot(
object,
assay_name = "counts",
genes = gene_spec(c(A = "GeneID:11185", B = "GeneID:10677")),
violin = TRUE
)