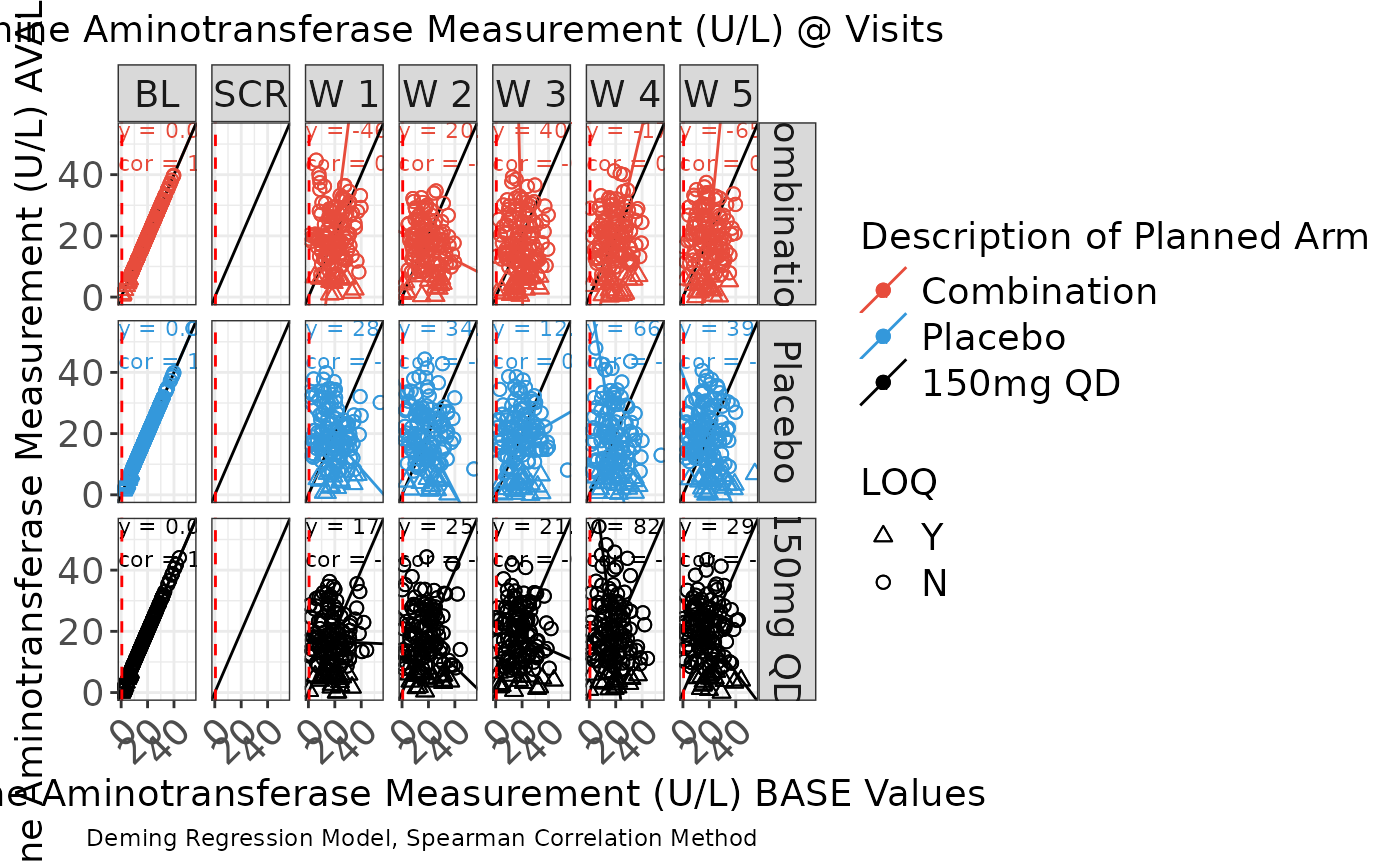
g_scatterplot() is deprecated. Please use
g_correlationplot() instead. Default plot displays scatter facetted by
visit with color attributed treatment arms and symbol attributed LOQ values.
Usage
g_scatterplot(
label = "Scatter Plot",
data,
param_var = "PARAMCD",
param = "CRP",
xaxis_var = "BASE",
yaxis_var = "AVAL",
trt_group = "ARM",
visit = "AVISITCD",
loq_flag_var = "LOQFL",
unit = "AVALU",
xlim = c(NA, NA),
ylim = c(NA, NA),
color_manual = NULL,
shape_manual = NULL,
facet_ncol = 2,
facet = FALSE,
facet_var = "ARM",
reg_line = FALSE,
hline = NULL,
vline = NULL,
rotate_xlab = FALSE,
font_size = 12,
dot_size = NULL,
reg_text_size = 3
)Arguments
- label
text string to used to identify plot.
- data
ADaMstructured analysis laboratory data frame e.g.ADLB.- param_var
name of variable containing biomarker codes e.g.
PARAMCD.- param
biomarker to visualize e.g.
IGG.- xaxis_var
name of variable containing biomarker results displayed on X-axis e.g.
BASE.- yaxis_var
name of variable containing biomarker results displayed on Y-axis e.g.
AVAL.- trt_group
name of variable representing treatment group e.g.
ARM.- visit
name of variable containing nominal visits e.g.
AVISITCD.- loq_flag_var
name of variable containing
LOQflag e.g.LOQFL.- unit
name of variable containing biomarker unit e.g.
AVALU.- xlim
('numeric vector') optional, a vector of length 2 to specify the minimum and maximum of the x-axis if the default limits are not suitable.
- ylim
('numeric vector') optional, a vector of length 2 to specify the minimum and maximum of the y-axis if the default limits are not suitable.
- color_manual
vector of colors applied to treatment values.
- shape_manual
vector of symbols applied to
LOQvalues.- facet_ncol
number of facets per row.
- facet
set layout to use treatment facetting.
- facet_var
variable to use for treatment facetting.
- reg_line
include regression line and annotations for slope and coefficient in visualization. Use with facet = TRUE.
- hline
y-axis value to position a horizontal line.
- vline
x-axis value to position a vertical line.
- rotate_xlab
45 degree rotation of x-axis label values.
- font_size
font size control for title, x-axis label, y-axis label and legend.
- dot_size
plot dot size.
- reg_text_size
font size control for regression line annotations.
Examples
# Example using ADaM structure analysis dataset.
library(stringr)
# original ARM value = dose value
arm_mapping <- list(
"A: Drug X" = "150mg QD", "B: Placebo" = "Placebo", "C: Combination" = "Combination"
)
color_manual <- c("150mg QD" = "#000000", "Placebo" = "#3498DB", "Combination" = "#E74C3C")
# assign LOQ flag symbols: circles for "N" and triangles for "Y", squares for "NA"
shape_manual <- c("N" = 1, "Y" = 2, "NA" = 0)
ADLB <- rADLB
var_labels <- lapply(ADLB, function(x) attributes(x)$label)
ADLB <- ADLB %>%
mutate(AVISITCD = case_when(
AVISIT == "SCREENING" ~ "SCR",
AVISIT == "BASELINE" ~ "BL",
grepl("WEEK", AVISIT) ~
paste(
"W",
trimws(
substr(
AVISIT,
start = 6,
stop = str_locate(AVISIT, "DAY") - 1
)
)
),
TRUE ~ NA_character_
)) %>%
mutate(AVISITCDN = case_when(
AVISITCD == "SCR" ~ -2,
AVISITCD == "BL" ~ 0,
grepl("W", AVISITCD) ~ as.numeric(gsub("\\D+", "", AVISITCD)),
TRUE ~ NA_real_
)) %>%
# use ARMCD values to order treatment in visualization legend
mutate(TRTORD = ifelse(grepl("C", ARMCD), 1,
ifelse(grepl("B", ARMCD), 2,
ifelse(grepl("A", ARMCD), 3, NA)
)
)) %>%
mutate(ARM = as.character(arm_mapping[match(ARM, names(arm_mapping))])) %>%
mutate(ARM = factor(ARM) %>%
reorder(TRTORD))
attr(ADLB[["ARM"]], "label") <- var_labels[["ARM"]]
g_scatterplot(
label = "Scatter Plot",
data = ADLB,
param_var = "PARAMCD",
param = c("ALT"),
xaxis_var = "BASE",
yaxis_var = "AVAL",
trt_group = "ARM",
visit = "AVISITCD",
loq_flag_var = "LOQFL",
unit = "AVALU",
color_manual = color_manual,
shape_manual = shape_manual,
facet_ncol = 2,
facet = TRUE,
facet_var = "ARM",
reg_line = TRUE,
hline = NULL,
vline = .5,
rotate_xlab = TRUE,
font_size = 14,
dot_size = 2,
reg_text_size = 3
)
#> Warning: `g_scatterplot()` was deprecated in goshawk 0.1.15.
#> ℹ You should use goshawk::g_correlationplot instead of goshawk::g_scatterplot