Function to create line plot of summary statistics over time.
Usage
g_lineplot(
label = "Line Plot",
data,
biomarker_var = "PARAMCD",
biomarker_var_label = "PARAM",
biomarker,
value_var = "AVAL",
unit_var = "AVALU",
loq_flag_var = "LOQFL",
ylim = c(NA, NA),
trt_group,
trt_group_level = NULL,
shape = NULL,
shape_type = NULL,
time,
time_level = NULL,
color_manual = NULL,
line_type = NULL,
median = FALSE,
hline_arb = numeric(0),
hline_arb_color = "red",
hline_arb_label = "Horizontal line",
xtick = ggplot2::waiver(),
xlabel = xtick,
rotate_xlab = FALSE,
plot_font_size = 12,
dodge = 0.4,
plot_height = 989,
count_threshold = 0,
table_font_size = 12,
display_center_tbl = TRUE
)Arguments
- label
text string to be displayed as plot label.
- data
ADaMstructured analysis laboratory data frame e.g.ADLB.- biomarker_var
name of variable containing biomarker names.
- biomarker_var_label
name of variable containing biomarker labels.
- biomarker
biomarker name to be analyzed.
- value_var
name of variable containing biomarker results.
- unit_var
name of variable containing biomarker result unit.
- loq_flag_var
name of variable containing
LOQflag e.g.LOQFL.- ylim
('numeric vector') optional, a vector of length 2 to specify the minimum and maximum of the y-axis if the default limits are not suitable.
- trt_group
name of variable representing treatment group.
- trt_group_level
vector that can be used to define the factor level of
trt_group.- shape
categorical variable whose levels are used to split the plot lines.
- shape_type
vector of symbol types.
- time
name of variable containing visit names.
- time_level
vector that can be used to define the factor level of time. Only use it when x-axis variable is character or factor.
- color_manual
vector of colors.
- line_type
vector of line types.
- median
boolean whether to display median results.
- hline_arb
('numeric vector') value identifying intercept for arbitrary horizontal lines.
- hline_arb_color
('character vector') optional, color for the arbitrary horizontal lines.
- hline_arb_label
('character vector') optional, label for the legend to the arbitrary horizontal lines.
- xtick
a vector to define the tick values of time in x-axis. Default value is
ggplot2::waiver().- xlabel
vector with same length of
xtickto define the label of x-axis tick values. Default value isggplot2::waiver().- rotate_xlab
boolean whether to rotate x-axis labels.
- plot_font_size
control font size for title, x-axis, y-axis and legend font.
- dodge
control position dodge.
- plot_height
height of produced plot. 989 pixels by default.
- count_threshold
integerminimum number observations needed to show the appropriate bar and point on the plot. Default: 0- table_font_size
floatcontrols the font size of the values printed in the table. Default: 12- display_center_tbl
boolean whether to include table of means or medians
Details
Currently, the output plot can display mean and median of input value. For mean, the error bar denotes 95\ quartile.
Examples
# Example using ADaM structure analysis dataset.
library(stringr)
library(dplyr)
library(nestcolor)
# original ARM value = dose value
arm_mapping <- list(
"A: Drug X" = "150mg QD", "B: Placebo" = "Placebo", "C: Combination" = "Combination"
)
color_manual <- c("150mg QD" = "thistle", "Placebo" = "orange", "Combination" = "steelblue")
type_manual <- c("150mg QD" = "solid", "Placebo" = "dashed", "Combination" = "dotted")
ADSL <- rADSL %>% filter(!(ARM == "B: Placebo" & AGE < 40))
ADLB <- rADLB
ADLB <- right_join(ADLB, ADSL[, c("STUDYID", "USUBJID")])
#> Joining with `by = join_by(STUDYID, USUBJID)`
var_labels <- lapply(ADLB, function(x) attributes(x)$label)
ADLB <- ADLB %>%
mutate(AVISITCD = case_when(
AVISIT == "SCREENING" ~ "SCR",
AVISIT == "BASELINE" ~ "BL",
grepl("WEEK", AVISIT) ~
paste(
"W",
trimws(
substr(
AVISIT,
start = 6,
stop = str_locate(AVISIT, "DAY") - 1
)
)
),
TRUE ~ NA_character_
)) %>%
mutate(AVISITCDN = case_when(
AVISITCD == "SCR" ~ -2,
AVISITCD == "BL" ~ 0,
grepl("W", AVISITCD) ~ as.numeric(gsub("\\D+", "", AVISITCD)),
TRUE ~ NA_real_
)) %>%
# use ARMCD values to order treatment in visualization legend
mutate(TRTORD = ifelse(grepl("C", ARMCD), 1,
ifelse(grepl("B", ARMCD), 2,
ifelse(grepl("A", ARMCD), 3, NA)
)
)) %>%
mutate(ARM = as.character(arm_mapping[match(ARM, names(arm_mapping))])) %>%
mutate(ARM = factor(ARM) %>%
reorder(TRTORD))
attr(ADLB[["ARM"]], "label") <- var_labels[["ARM"]]
g_lineplot(
label = "Line Plot",
data = ADLB,
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = NULL,
time = "AVISITCDN",
color_manual = color_manual,
line_type = type_manual,
median = FALSE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c(0, 1, 5),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 600
)
 g_lineplot(
label = "Line Plot",
data = ADLB,
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = NULL,
time = "AVISITCD",
color_manual = NULL,
line_type = type_manual,
median = TRUE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c("BL", "W 1", "W 5"),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 600
)
g_lineplot(
label = "Line Plot",
data = ADLB,
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = NULL,
time = "AVISITCD",
color_manual = NULL,
line_type = type_manual,
median = TRUE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c("BL", "W 1", "W 5"),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 600
)
 g_lineplot(
label = "Line Plot",
data = ADLB,
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = NULL,
time = "AVISITCD",
color_manual = color_manual,
line_type = type_manual,
median = FALSE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c("BL", "W 1", "W 5"),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 600,
count_threshold = 90,
table_font_size = 15
)
g_lineplot(
label = "Line Plot",
data = ADLB,
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = NULL,
time = "AVISITCD",
color_manual = color_manual,
line_type = type_manual,
median = FALSE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c("BL", "W 1", "W 5"),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 600,
count_threshold = 90,
table_font_size = 15
)
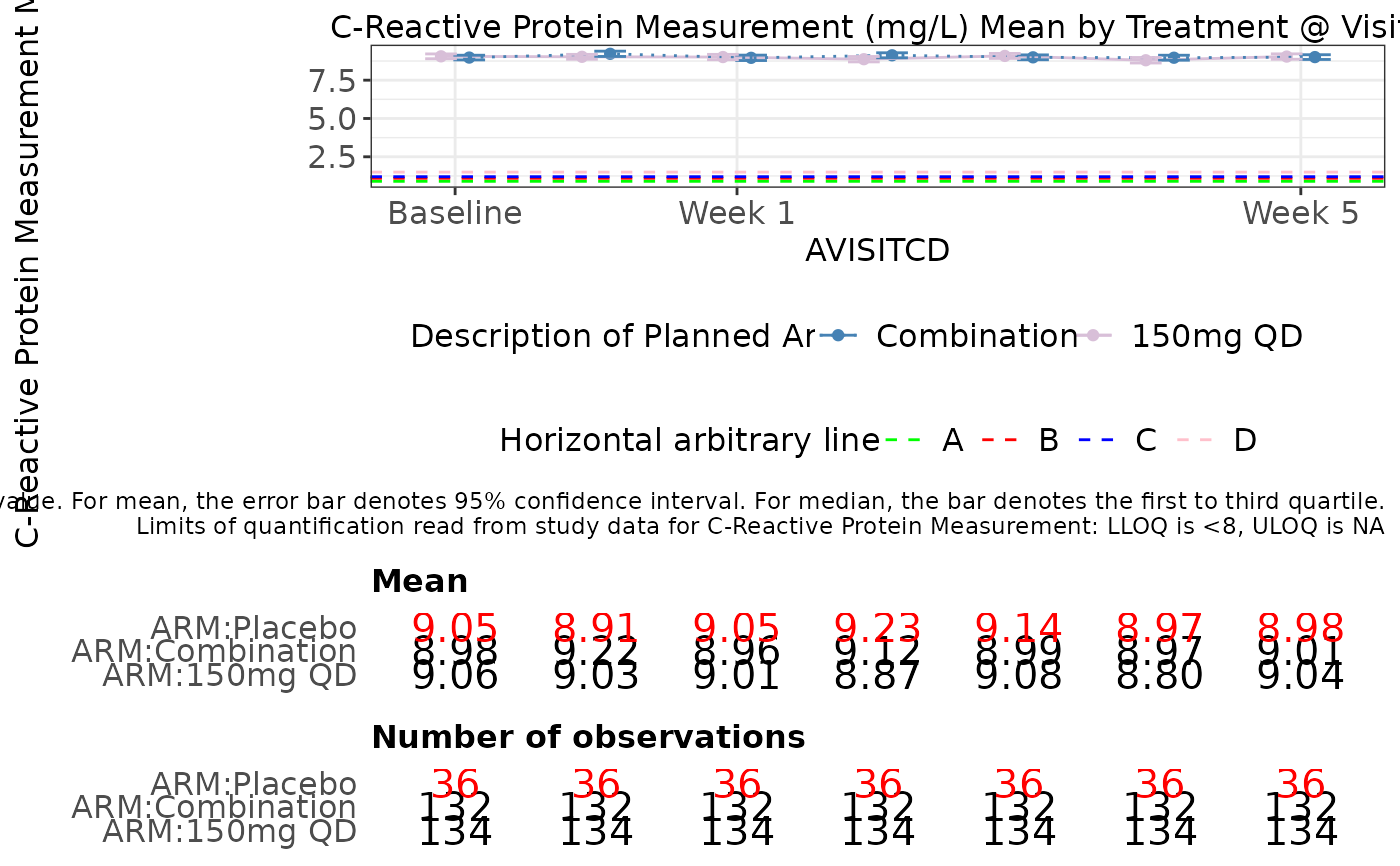 g_lineplot(
label = "Line Plot",
data = ADLB,
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = NULL,
time = "AVISITCDN",
color_manual = color_manual,
line_type = type_manual,
median = TRUE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c(0, 1, 5),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 600
)
g_lineplot(
label = "Line Plot",
data = ADLB,
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = NULL,
time = "AVISITCDN",
color_manual = color_manual,
line_type = type_manual,
median = TRUE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c(0, 1, 5),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 600
)
 g_lineplot(
label = "Line Plot",
data = subset(ADLB, SEX %in% c("M", "F")),
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = "SEX",
time = "AVISITCDN",
color_manual = color_manual,
line_type = type_manual,
median = FALSE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c(0, 1, 5),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 1500
)
g_lineplot(
label = "Line Plot",
data = subset(ADLB, SEX %in% c("M", "F")),
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = "SEX",
time = "AVISITCDN",
color_manual = color_manual,
line_type = type_manual,
median = FALSE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c(0, 1, 5),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 1500
)
 g_lineplot(
label = "Line Plot",
data = subset(ADLB, SEX %in% c("M", "F")),
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = "SEX",
time = "AVISITCDN",
color_manual = NULL,
median = FALSE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c(0, 1, 5),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 1500
)
g_lineplot(
label = "Line Plot",
data = subset(ADLB, SEX %in% c("M", "F")),
biomarker_var = "PARAMCD",
biomarker = "CRP",
value_var = "AVAL",
trt_group = "ARM",
shape = "SEX",
time = "AVISITCDN",
color_manual = NULL,
median = FALSE,
hline_arb = c(.9, 1.1, 1.2, 1.5),
hline_arb_color = c("green", "red", "blue", "pink"),
hline_arb_label = c("A", "B", "C", "D"),
xtick = c(0, 1, 5),
xlabel = c("Baseline", "Week 1", "Week 5"),
rotate_xlab = FALSE,
plot_height = 1500
)
